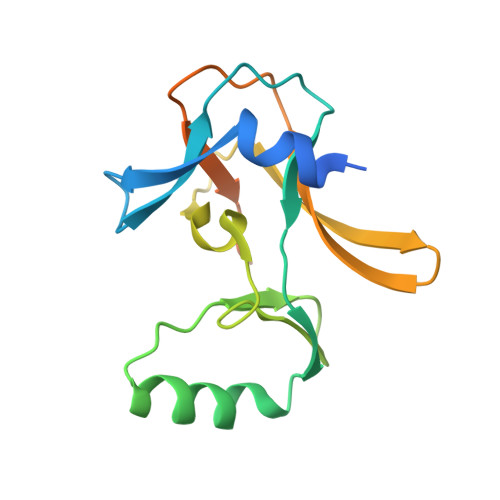
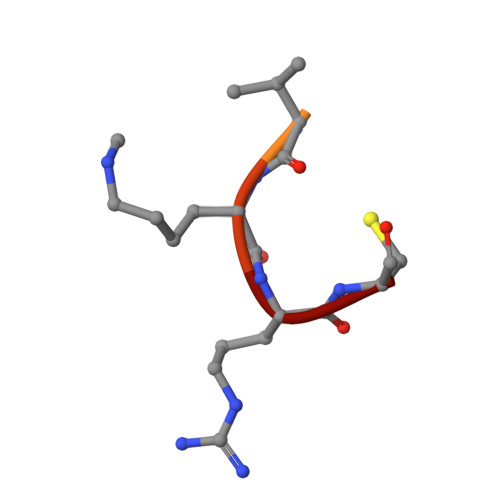
Non-canonical MLL1 activity regulates centromeric phase separation and genome stability.
Sha, L., Yang, Z., An, S., Yang, W., Kim, S., Oh, H., Xu, J., Yin, J., Wang, H., Lenz, H.J., An, W., Cho, U.S., Dou, Y.(2023) Nat Cell Biol 25: 1637-1649
- PubMed: 37945831
- DOI: https://doi.org/10.1038/s41556-023-01270-1
- Primary Citation of Related Structures:
7U5V - PubMed Abstract:
Epigenetic dysregulation is a prominent feature in cancer, as exemplified by frequent mutations in chromatin regulators, including the MLL/KMT2 family of histone methyltransferases. Although MLL1/KMT2A activity on H3K4 methylation is well documented, their non-canonical activities remain mostly unexplored. Here we show that MLL1/KMT2A methylates Borealin K143 in the intrinsically disordered region essential for liquid-liquid phase separation of the chromosome passenger complex (CPC). The co-crystal structure highlights the distinct binding mode of the MLL1 SET domain with Borealin K143. Inhibiting MLL1 activity or mutating Borealin K143 to arginine perturbs CPC phase separation, reduces Aurora kinase B activity, and impairs the resolution of erroneous kinetochore-microtubule attachments and sister-chromatid cohesion. They significantly increase chromosome instability and aneuploidy in a subset of hepatocellular carcinoma, resulting in growth inhibition. These results demonstrate a non-redundant function of MLL1 in regulating inner centromere liquid condensates and genome stability via a non-canonical enzymatic activity.
- Department of Medicine, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA.
Organizational Affiliation: