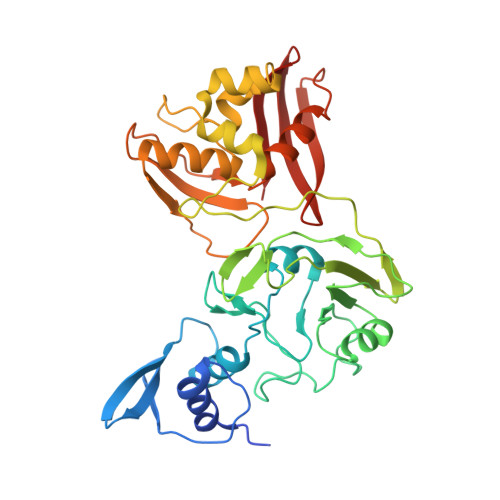
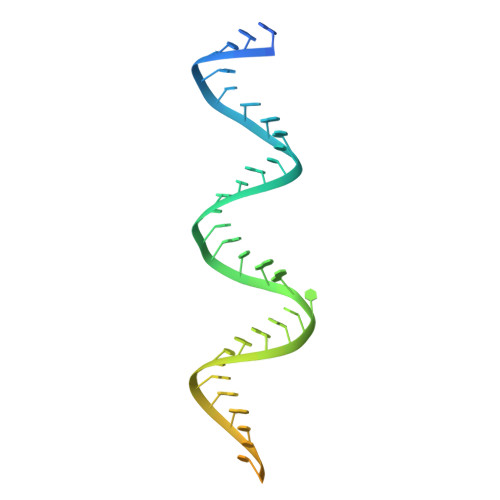
Flipped over U: structural basis for dsRNA cleavage by the SARS-CoV-2 endoribonuclease.
Frazier, M.N., Wilson, I.M., Krahn, J.M., Butay, K.J., Dillard, L.B., Borgnia, M.J., Stanley, R.E.(2022) Nucleic Acids Res 50: 8290-8301
- PubMed: 35801916
- DOI: https://doi.org/10.1093/nar/gkac589
- Primary Citation of Related Structures:
7TJ2, 7TQV - PubMed Abstract:
Coronaviruses generate double-stranded (ds) RNA intermediates during viral replication that can activate host immune sensors. To evade activation of the host pattern recognition receptor MDA5, coronaviruses employ Nsp15, which is a uridine-specific endoribonuclease. Nsp15 is proposed to associate with the coronavirus replication-transcription complex within double-membrane vesicles to cleave these dsRNA intermediates. How Nsp15 recognizes and processes dsRNA is poorly understood because previous structural studies of Nsp15 have been limited to small single-stranded (ss) RNA substrates. Here we present cryo-EM structures of SARS-CoV-2 Nsp15 bound to a 52nt dsRNA. We observed that the Nsp15 hexamer forms a platform for engaging dsRNA across multiple protomers. The structures, along with site-directed mutagenesis and RNA cleavage assays revealed critical insight into dsRNA recognition and processing. To process dsRNA Nsp15 utilizes a base-flipping mechanism to properly orient the uridine within the active site for cleavage. Our findings show that Nsp15 is a distinctive endoribonuclease that can cleave both ss- and dsRNA effectively.
- Signal Transduction Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Department of Health and Human Services, 111 T. W. Alexander Drive, Research Triangle Park, NC 27709, USA.
Organizational Affiliation: