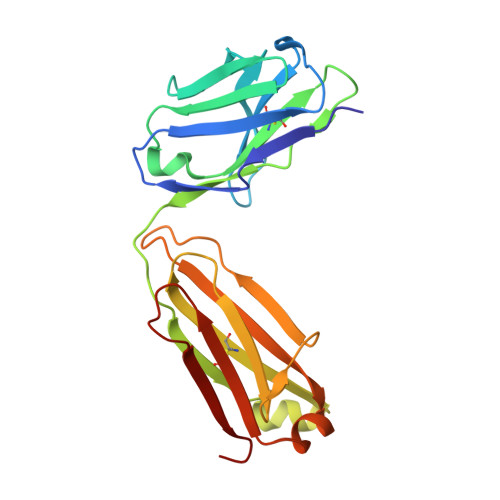
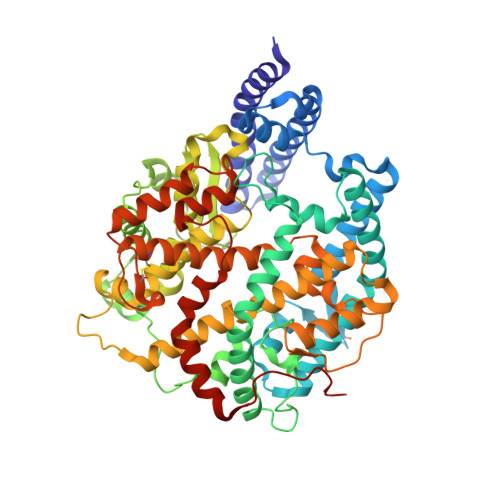
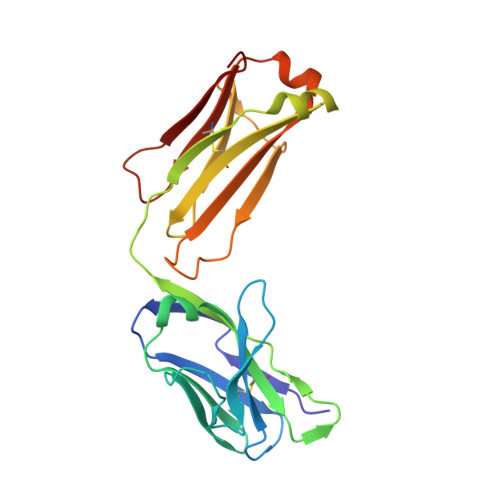
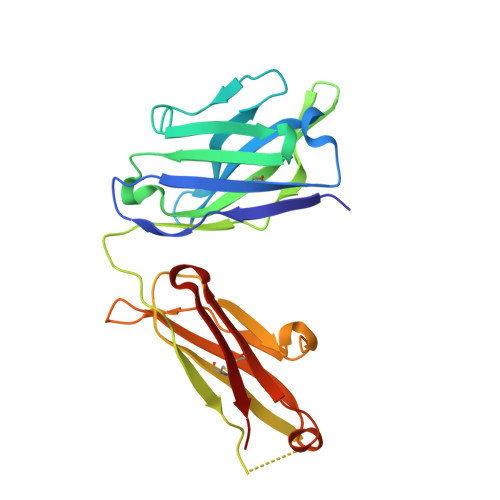
Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement.
McCallum, M., Czudnochowski, N., Rosen, L.E., Zepeda, S.K., Bowen, J.E., Walls, A.C., Hauser, K., Joshi, A., Stewart, C., Dillen, J.R., Powell, A.E., Croll, T.I., Nix, J., Virgin, H.W., Corti, D., Snell, G., Veesler, D.(2022) Science 375: 864-868
- PubMed: 35076256
- DOI: https://doi.org/10.1126/science.abn8652
- Primary Citation of Related Structures:
7TLY, 7TLZ, 7TM0, 7TN0 - PubMed Abstract:
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant of concern evades antibody-mediated immunity that comes from vaccination or infection with earlier variants due to accumulation of numerous spike mutations. To understand the Omicron antigenic shift, we determined cryo-electron microscopy and x-ray crystal structures of the spike protein and the receptor-binding domain bound to the broadly neutralizing sarbecovirus monoclonal antibody (mAb) S309 (the parent mAb of sotrovimab) and to the human ACE2 receptor. We provide a blueprint for understanding the marked reduction of binding of other therapeutic mAbs that leads to dampened neutralizing activity. Remodeling of interactions between the Omicron receptor-binding domain and human ACE2 likely explains the enhanced affinity for the host receptor relative to the ancestral virus.
- Department of Biochemistry, University of Washington, Seattle, WA 98195, USA.
Organizational Affiliation: