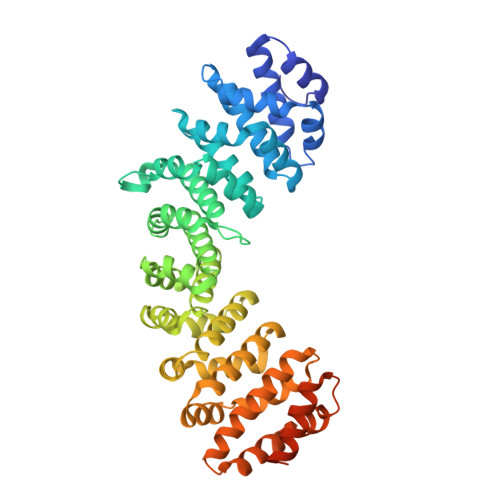
Structural basis of nuclear transport for NEIL DNA glycosylases mediated by importin-alpha.
Moraes, I.R., de Oliveira, H.C., Fontes, M.R.M.(2023) Biochim Biophys Acta Proteins Proteom 1872: 140974-140974
- PubMed: 38065227
- DOI: https://doi.org/10.1016/j.bbapap.2023.140974
- Primary Citation of Related Structures:
7TMX, 7TMY - PubMed Abstract:
NEIL glycosylases, including NEIL1, NEIL2, and NEIL3, play a crucial role in the base excision DNA repair pathway (BER). The classical importin pathway mediated by importin α/β and cargo proteins containing nuclear localization sequences (NLS) is the most common transport mechanism of DNA repair proteins to the nucleus. Previous studies have identified putative NLSs located at the C-terminus of NEIL3 and NEIL1. Crystallographic, bioinformatics, calorimetric (ITC), and fluorescence assays were used to investigate the interaction between NEIL1 and NEIL3 putative NLSs and importin-α (Impα). Our findings showed that NEIL3 contains a typical cNLS, with medium affinity for the major binding site of Impα. In contrast, crystallographic analysis of NEIL1 NLS revealed its binding to Impα, but with high B-factors and a lack of electron density at the linker region. ITC and fluorescence assays indicated no detectable affinity between NEIL1 NLS and Impα. These data suggest that NEIL1 NLS is a non-classical NLS with low affinity to Impα. Additionally, we compared the binding mode of NEIL3 and NEIL1 with Mus musculus Impα to human isoforms HsImpα1 and HsImpα3, which revealed interesting binding differences for HsImpα3 variant. NEIL3 is a classical medium affinity monopartite NLS, while NEIL1 is likely to be an unclassical low-affinity bipartite NLS. The base excision repair pathway is one of the primary systems involved in repairing DNA. Thus, understanding the mechanisms of nuclear transport of NEIL proteins is crucial for comprehending the role of these proteins in DNA repair and disease development.
- Departamento de Biofísica e Farmacologia, Instituto de Biociências, Universidade Estadual Paulista (UNESP), Botucatu, SP, Brazil.
Organizational Affiliation: