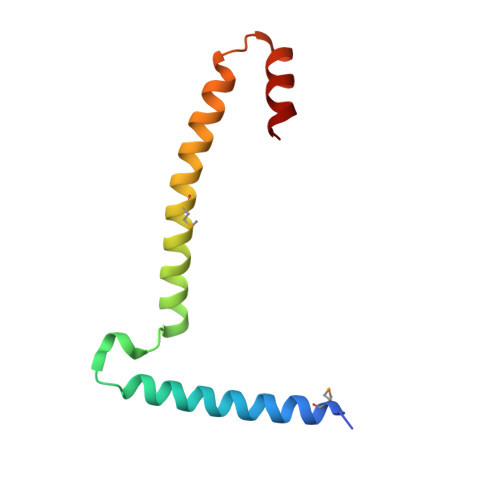
Structural basis of higher order oligomerization of KSHV inhibitor of cGAS.
Bhowmik, D., Tian, Y., Wang, B., Zhu, F., Yin, Q.(2022) Proc Natl Acad Sci U S A 119: e2200285119-e2200285119
- PubMed: 35939686
- DOI: https://doi.org/10.1073/pnas.2200285119
- Primary Citation of Related Structures:
7TDQ - PubMed Abstract:
Kaposi's sarcoma-associated herpesvirus (KSHV) inhibitor of cyclic GMP-AMP synthase (cGAS) (KicGAS) encoded by ORF52 is a conserved major tegument protein of KSHV and the first reported viral inhibitor of cGAS. In our previous study, we found that KicGAS is highly oligomerized in solution and that oligomerization is required for its cooperative DNA binding and for inhibiting DNA-induced phase separation and activation of cGAS. However, how KicGAS oligomerizes remained unclear. Here, we present the crystal structure of KicGAS at 2.5 Å resolution, which reveals an "L"-shaped molecule with each arm of the L essentially formed by a single α helix (α1 and α2). Antiparallel dimerization of α2 helices from two KicGAS molecules leads to a unique "Z"-shaped dimer. Surprisingly, α1 is also a dimerization domain. It forms a parallel dimeric leucine zipper with the α1 from a neighboring dimer, leading to the formation of an infinite chain of KicGAS dimers. Residues involved in leucine zipper dimer formation are among the most conserved residues across ORF52 homologs of gammaherpesviruses. The self-oligomerization increases the valence and cooperativity of interaction with DNA. The resultant multivalent interaction is critical for the formation of liquid condensates with DNA and consequent sequestration of DNA from being sensed by cGAS, explaining its role in restricting cGAS activation. The structure presented here not only provides a mechanistic understanding of the function of KicGAS but also informs a molecular target for rational design of antivirals against KSHV and related viruses.
- Department of Biological Science, Florida State University, Tallahassee, FL 32306.
Organizational Affiliation: