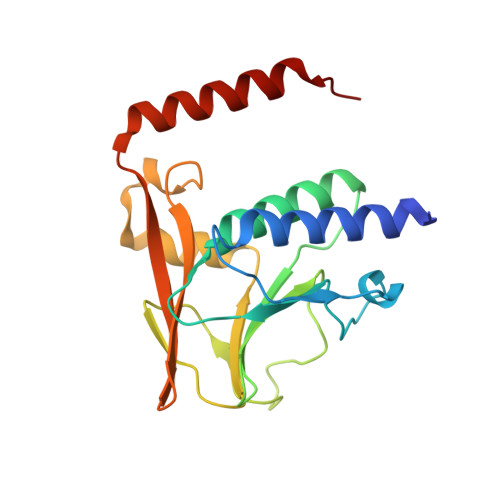
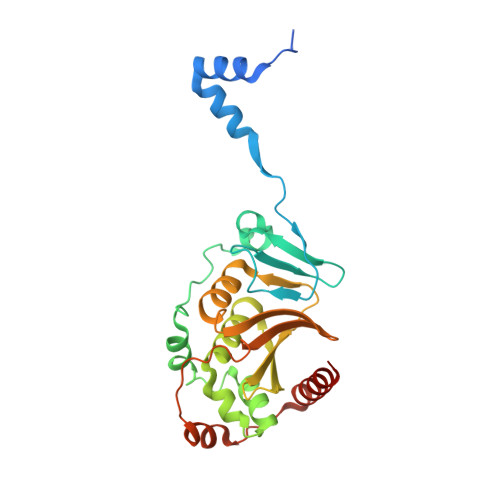
The nuclear egress complex of Epstein-Barr virus buds membranes through an oligomerization-driven mechanism.
Thorsen, M.K., Draganova, E.B., Heldwein, E.E.(2022) PLoS Pathog 18: e1010623-e1010623
- PubMed: 35802751
- DOI: https://doi.org/10.1371/journal.ppat.1010623
- Primary Citation of Related Structures:
7T7I - PubMed Abstract:
During replication, herpesviral capsids are translocated from the nucleus into the cytoplasm by an unusual mechanism, termed nuclear egress, that involves capsid budding at the inner nuclear membrane. This process is mediated by the viral nuclear egress complex (NEC) that deforms the membrane around the capsid. Although the NEC is essential for capsid nuclear egress across all three subfamilies of the Herpesviridae, most studies to date have focused on the NEC homologs from alpha- and beta- but not gammaherpesviruses. Here, we report the crystal structure of the NEC from Epstein-Barr virus (EBV), a prototypical gammaherpesvirus. The structure resembles known structures of NEC homologs yet is conformationally dynamic. We also show that purified, recombinant EBV NEC buds synthetic membranes in vitro and forms membrane-bound coats of unknown geometry. However, unlike other NEC homologs, EBV NEC forms dimers in the crystals instead of hexamers. The dimeric interfaces observed in the EBV NEC crystals are similar to the hexameric interfaces observed in other NEC homologs. Moreover, mutations engineered to disrupt the dimeric interface reduce budding. Putting together these data, we propose that EBV NEC-mediated budding is driven by oligomerization into membrane-bound coats.
- Department of Molecular Biology and Microbiology, Tufts University School of Medicine, Boston, Massachusetts, United States of America.
Organizational Affiliation: