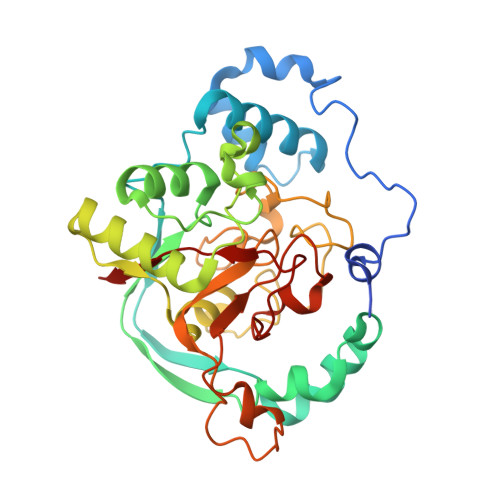
Crystal Structure, Modeling, and Identification of Key Residues Provide Insights into the Mechanism of the Key Toxoflavin Biosynthesis Protein ToxD.
Justen, S.F., Fenwick, M.K., Axt, K.K., Cherry, J.A., Ealick, S.E., Philmus, B.(2025) Biochemistry 64: 1199-1211
- PubMed: 40047534
- DOI: https://doi.org/10.1021/acs.biochem.4c00421
- Primary Citation of Related Structures:
7SPQ - PubMed Abstract:
Toxoflavin, a toxic secondary metabolite produced by a variety of bacteria, has been implicated as a causative agent in food poisoning and a virulence factor in phytopathogenic bacteria. This toxin is produced by genes encoded in the tox operon in Burkholderia glumae , in which the encoded protein, ToxD, was previously characterized as essential for toxoflavin production. To better understand the function of ToxD in toxoflavin biosynthesis and provide a basis for future work to develop inhibitors of ToxD, we undertook the identification of structurally and catalytically important amino acid residues through a combination of X-ray crystallography and site directed mutagenesis. We solved the structure of Bg ToxD, which crystallized as a dimer, to 1.8 Å resolution. We identified a citrate molecule in the putative active site. To investigate the role of individual residues, we used Pseudomonas protegens Pf-5, a BL1 plant protective bacterium known to produce toxoflavin, and created mutants in the ToxD-homologue PFL1035. Using a multiple sequence alignment and the Bg ToxD structure, we identified and explored the functional importance of 12 conserved residues in the putative active site. Eight variants of PFL1035 resulted in no observable production of toxoflavin. In contrast, four ToxD variants resulted in reduced but detectable toxoflavin production suggesting a nonessential role. The crystal structure and structural models of the substrate and intermediate bound enzyme provide a molecular interpretation for the mutagenesis data.
- Department of Pharmaceutical Sciences, Oregon State University, Corvallis, Oregon 97331, United States.
Organizational Affiliation: