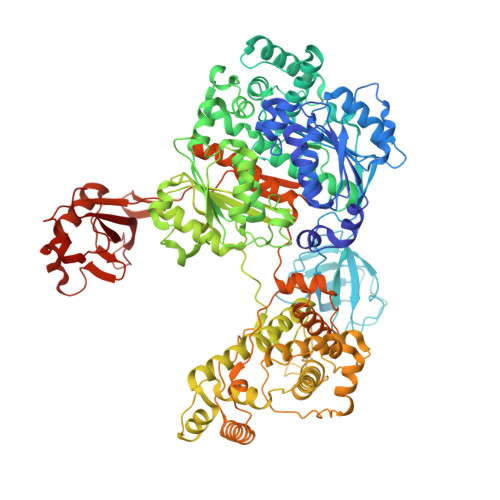
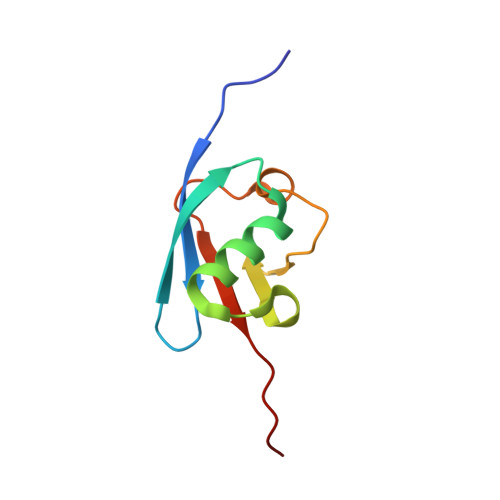
Crystal structures reveal catalytic and regulatory mechanisms of the dual-specificity ubiquitin/FAT10 E1 enzyme Uba6.
Yuan, L., Gao, F., Lv, Z., Nayak, D., Nayak, A., Santos Bury, P.D., Cano, K.E., Jia, L., Oleinik, N., Atilgan, F.C., Ogretmen, B., Williams, K.M., Davies, C., El Oualid, F., Wasmuth, E.V., Olsen, S.K.(2022) Nat Commun 13: 4880-4880
- PubMed: 35986001
- DOI: https://doi.org/10.1038/s41467-022-32613-5
- Primary Citation of Related Structures:
7SOL - PubMed Abstract:
The E1 enzyme Uba6 initiates signal transduction by activating ubiquitin and the ubiquitin-like protein FAT10 in a two-step process involving sequential catalysis of adenylation and thioester bond formation. To gain mechanistic insights into these processes, we determined the crystal structure of a human Uba6/ubiquitin complex. Two distinct architectures of the complex are observed: one in which Uba6 adopts an open conformation with the active site configured for catalysis of adenylation, and a second drastically different closed conformation in which the adenylation active site is disassembled and reconfigured for catalysis of thioester bond formation. Surprisingly, an inositol hexakisphosphate (InsP6) molecule binds to a previously unidentified allosteric site on Uba6. Our structural, biochemical, and biophysical data indicate that InsP6 allosterically inhibits Uba6 activity by altering interconversion of the open and closed conformations of Uba6 while also enhancing its stability. In addition to revealing the molecular mechanisms of catalysis by Uba6 and allosteric regulation of its activities, our structures provide a framework for developing Uba6-specific inhibitors and raise the possibility of allosteric regulation of other E1s by naturally occurring cellular metabolites.
- Department of Biochemistry & Structural Biology, University of Texas Health Science Center at San Antonio, San Antonio, TX, 78229, USA.
Organizational Affiliation: