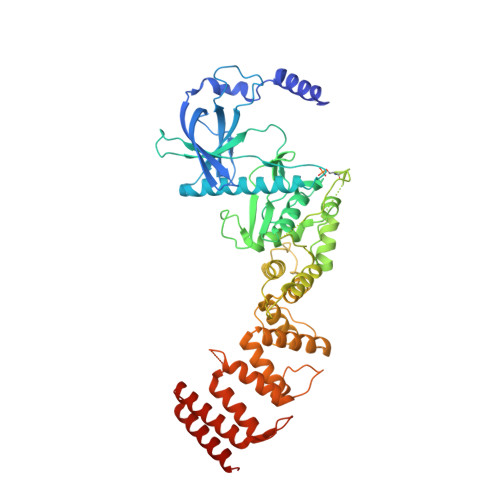
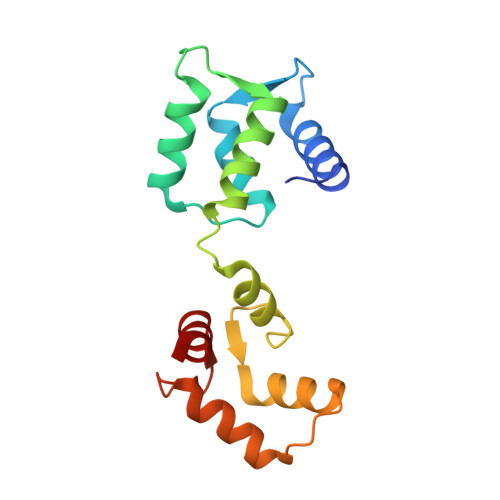
Structural basis for the calmodulin-mediated activation of eukaryotic elongation factor 2 kinase.
Piserchio, A., Isiorho, E.A., Long, K., Bohanon, A.L., Kumar, E.A., Will, N., Jeruzalmi, D., Dalby, K.N., Ghose, R.(2022) Sci Adv 8: eabo2039-eabo2039
- PubMed: 35857468
- DOI: https://doi.org/10.1126/sciadv.abo2039
- Primary Citation of Related Structures:
7SHQ - PubMed Abstract:
Translation is a tightly regulated process that ensures optimal protein quality and enables adaptation to energy/nutrient availability. The α-kinase eukaryotic elongation factor 2 kinase (eEF-2K), a key regulator of translation, specifically phosphorylates the guanosine triphosphatase eEF-2, thereby reducing its affinity for the ribosome and suppressing the elongation phase of protein synthesis. eEF-2K activation requires calmodulin binding and autophosphorylation at the primary stimulatory site, T348. Biochemical studies predict a calmodulin-mediated activation mechanism for eEF-2K distinct from other calmodulin-dependent kinases. Here, we resolve the atomic details of this mechanism through a 2.3-Å crystal structure of the heterodimeric complex of calmodulin and the functional core of eEF-2K (eEF-2K TR ). This structure, which represents the activated T348-phosphorylated state of eEF-2K TR , highlights an intimate association of the kinase with the calmodulin C-lobe, creating an "activation spine" that connects its amino-terminal calmodulin-targeting motif to its active site through a conserved regulatory element.
- Department of Chemistry and Biochemistry, The City College of New York, New York, NY 10031, USA.
Organizational Affiliation: