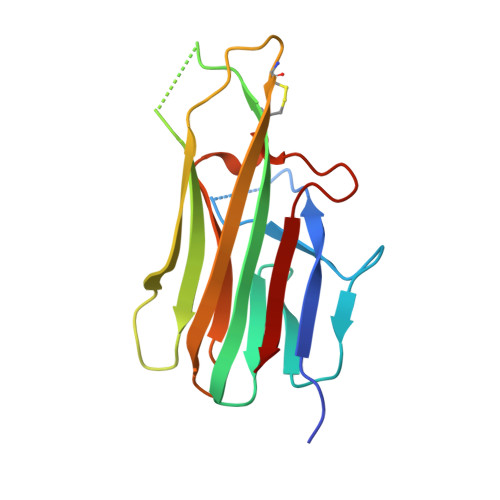
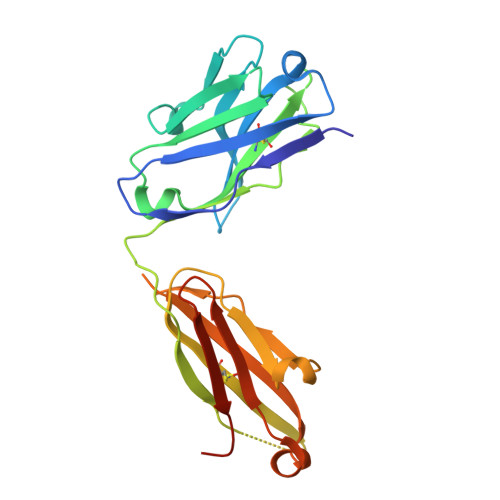
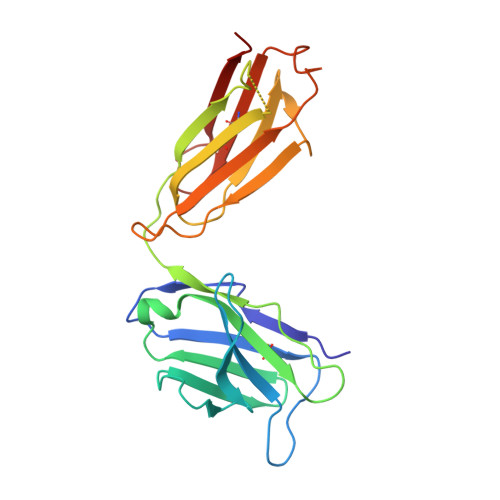
Structural Basis for Blocked Excited State Proton Transfer in a Fluorescent, Photoacidic Non-Canonical Amino Acid-Containing Antibody Fragment.
Henderson, J.N., Simmons, C.R., Mills, J.H.(2022) J Mol Biology 434: 167455-167455
- PubMed: 35033559
- DOI: https://doi.org/10.1016/j.jmb.2022.167455
- Primary Citation of Related Structures:
7SEN, 7SGM - PubMed Abstract:
The fluorescent non-canonical amino acid (fNCAA) L-(7-hydroxycoumarin-4-yl)ethylglycine (7-HCAA) contains a photoacidic 7-hydroxycoumarin (7-HC) side chain whose fluorescence properties can be tuned by its environment. In proteins, many alterations to 7-HCAA's fluorescence spectra have been reported including increases and decreases in intensity and red- and blue-shifted emission maxima. The ability to rationally design protein environments that alter 7-HCAA's fluorescence properties in predictable ways could lead to novel protein-based sensors of biological function. However, these efforts are likely limited by a lack of structural characterization of 7-HCAA-containing proteins. Here, we report the steady-state spectroscopic and x-ray crystallographic characterization of a 7-HCAA-containing antibody fragment (in the apo and antigen-bound forms) in which a substantially blue-shifted 7-HCAA emission maximum (∼70 nm) is observed relative to the free amino acid. Our structural characterization of these proteins provides evidence that the blue shift is a consequence of the fact that excited state proton transfer (ESPT) from the 7-HC phenol has been almost completely blocked by interactions with the protein backbone. Furthermore, a direct interaction between a residue in the antigen and the fluorophore served to further block proton transfer relative to the apoprotein. The structural basis of the unprecedented blue shift in 7-HCAA emission reported here provides a framework for the development of new fluorescent protein-based sensors.
- The Biodesign Center for Molecular Design and Biomimetics, Arizona State University, Tempe, AZ, USA.
Organizational Affiliation: