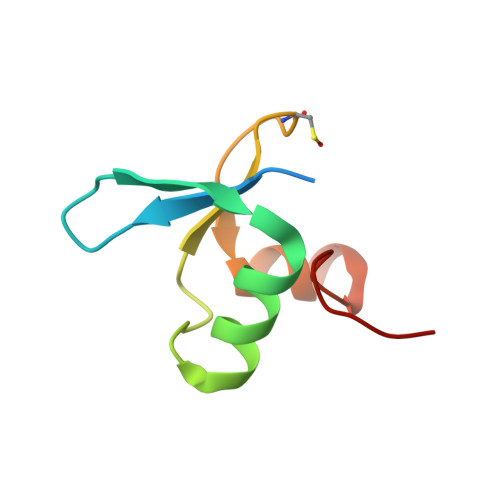
Molecular basis for GIGYF-TNRC6 complex assembly.
Sobti, M., Mead, B.J., Stewart, A.G., Igreja, C., Christie, M.(2023) RNA 29: 724-734
- PubMed: 36854607
- DOI: https://doi.org/10.1261/rna.079596.123
- Primary Citation of Related Structures:
7RUP, 7RUQ - PubMed Abstract:
The GIGYF proteins interact with 4EHP and RNA-associated proteins to elicit transcript-specific translational repression. However, the mechanism by which the GIGYF1/2-4EHP complex is recruited to its target transcripts remain unclear. Here, we report the crystal structures of the GYF domains from GIGYF1 and GIGYF2 in complex with proline-rich sequences from the miRISC-binding proteins TNRC6C and TNRC6A, respectively. The TNRC6 proline-rich motifs bind to a conserved array of aromatic residues on the surface of the GIGYF1/2 GYF domains, thereby bridging 4EHP to Argonaute-miRNA complexes. Our structures also reveal a phenylalanine residue conserved from yeast to human GYF domains that contributes to GIGYF2 thermostability. The molecular details we outline here are likely to be conserved between GIGYF1/2 and other RNA-binding proteins to elicit 4EHP-mediated repression in different biological contexts.
- Molecular, Structural and Computational Biology Division, The Victor Chang Cardiac Research Institute, Sydney, New South Wales 2010, Australia.
Organizational Affiliation: