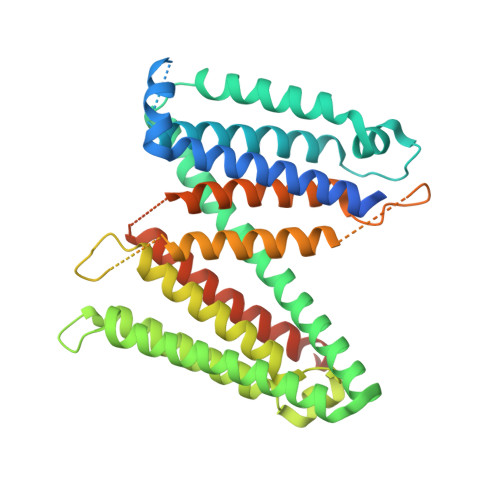
Antiviral HIV-1 SERINC restriction factors disrupt virus membrane asymmetry.
Leonhardt, S.A., Purdy, M.D., Grover, J.R., Yang, Z., Poulos, S., McIntire, W.E., Tatham, E.A., Erramilli, S.K., Nosol, K., Lai, K.K., Ding, S., Lu, M., Uchil, P.D., Finzi, A., Rein, A., Kossiakoff, A.A., Mothes, W., Yeager, M.(2023) Nat Commun 14: 4368-4368
- PubMed: 37474505
- DOI: https://doi.org/10.1038/s41467-023-39262-2
- Primary Citation of Related Structures:
7RU6, 7RUG - PubMed Abstract:
The host proteins SERINC3 and SERINC5 are HIV-1 restriction factors that reduce infectivity when incorporated into the viral envelope. The HIV-1 accessory protein Nef abrogates incorporation of SERINCs via binding to intracellular loop 4 (ICL4). Here, we determine cryoEM maps of full-length human SERINC3 and an ICL4 deletion construct, which reveal that hSERINC3 is comprised of two α-helical bundles connected by a ~ 40-residue, highly tilted, "crossmember" helix. The design resembles non-ATP-dependent lipid transporters. Consistently, purified hSERINCs reconstituted into proteoliposomes induce flipping of phosphatidylserine (PS), phosphatidylethanolamine and phosphatidylcholine. Furthermore, SERINC3, SERINC5 and the scramblase TMEM16F expose PS on the surface of HIV-1 and reduce infectivity, with similar results in MLV. SERINC effects in HIV-1 and MLV are counteracted by Nef and GlycoGag, respectively. Our results demonstrate that SERINCs are membrane transporters that flip lipids, resulting in a loss of membrane asymmetry that is strongly correlated with changes in Env conformation and loss of infectivity.
- The Phillip and Patricia Frost Institute for Chemistry and Molecular Science, University of Miami, Coral Gables, FL, 33146, USA.
Organizational Affiliation: