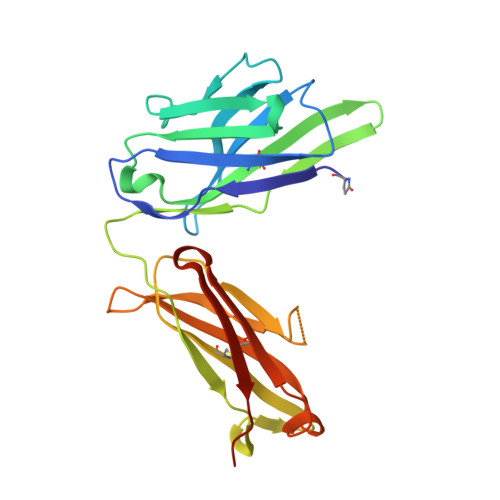
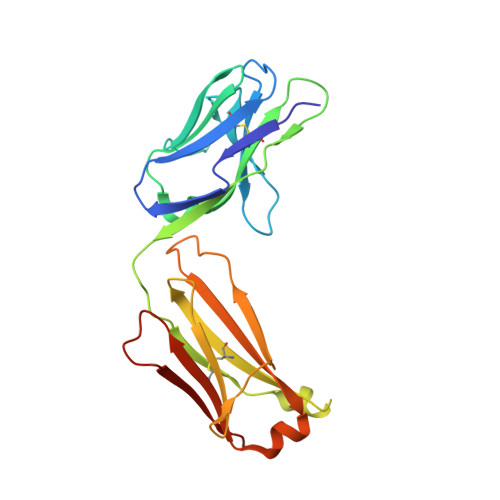
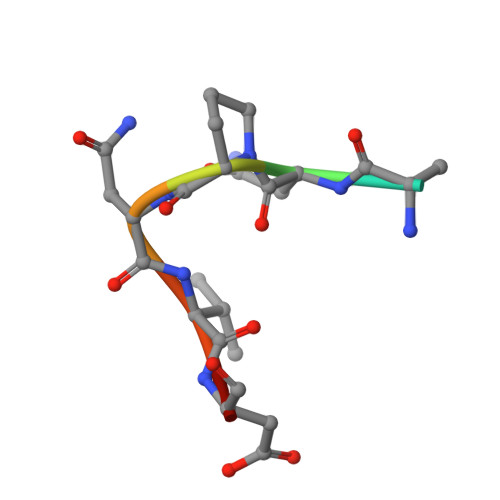
The light chain of the L9 antibody is critical for binding circumsporozoite protein minor repeats and preventing malaria.
Wang, L.T., Hurlburt, N.K., Schon, A., Flynn, B.J., Flores-Garcia, Y., Pereira, L.S., Kiyuka, P.K., Dillon, M., Bonilla, B., Zavala, F., Idris, A.H., Francica, J.R., Pancera, M., Seder, R.A.(2022) Cell Rep 38: 110367-110367
- PubMed: 35172158
- DOI: https://doi.org/10.1016/j.celrep.2022.110367
- Primary Citation of Related Structures:
7RQP, 7RQQ, 7RQR - PubMed Abstract:
L9 is a potent human monoclonal antibody (mAb) that preferentially binds two adjacent NVDP minor repeats and cross-reacts with NANP major repeats of the Plasmodium falciparum circumsporozoite protein (PfCSP) on malaria-infective sporozoites. Understanding this mAb's ontogeny and mechanisms of binding PfCSP will facilitate vaccine development. Here, we isolate mAbs clonally related to L9 and show that this B cell lineage has baseline NVDP affinity and evolves to acquire NANP reactivity. Pairing the L9 kappa light chain (L9κ) with clonally related heavy chains results in chimeric mAbs that cross-link two NVDPs, cross-react with NANP, and more potently neutralize sporozoites in vivo compared with their original light chain. Structural analyses reveal that the chimeric mAbs bound minor repeats in a type-1 β-turn seen in other repeat-specific antibodies. These data highlight the importance of L9κ in binding NVDP on PfCSP to neutralize sporozoites and suggest that PfCSP-based immunogens might be improved by presenting ≥2 NVDPs.
- Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: