The molecular basis for the development of adult T-cell leukemia/lymphoma in patients with an IRF4 K59R mutation.
Sundararaj, S., Seneviratne, S., Williams, S.J., Enders, A., Casarotto, M.G.(2022) Protein Sci 31: 787-796
- PubMed: 34913532
- DOI: https://doi.org/10.1002/pro.4260
- Primary Citation of Related Structures:
7RH2 - PubMed Abstract:
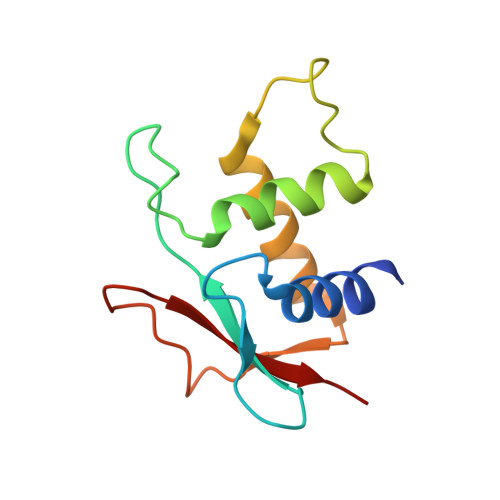
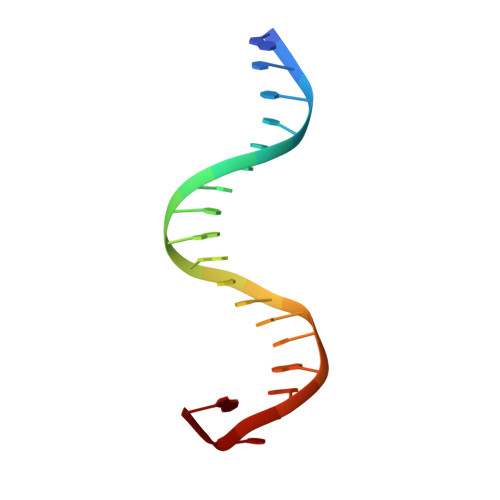
Interferon regulatory factor 4 (IRF4) is an essential regulator in the development of many immune cells, including B- and T-cells and has been implicated directly in numerous hematological malignancies, including adult T-cell leukemia/lymphoma (ATLL). Recently, an activating mutation in the DNA-binding domain of IRF4 (IRF4 K59R ) was found as a recurrent somatic mutation in ATLL patients. However, it remains unknown how this mutation gives rise to the observed oncogenic effect. To understand the mode of IRF4 K59R -mediated gain of function in ATLL pathogenesis, we have determined the structural and affinity basis of IRF4 K59R /DNA homodimer complex using X-ray crystallography and surface plasmon resonance. Our study shows that arginine substitution (R59) results in the reorientation of the side chain, enabling the guanidium group to interact with the phosphate backbone of the DNA helix. This markedly contrasts with the IRF4 WT wherein the K59 interacts exclusively with DNA bases. Further, the arginine mutation causes enhanced DNA bending, enabling the IRF4 K59R to interact more robustly with known DNA targets, as evidenced by increased binding affinity of the protein-DNA complex. Together, we demonstrate how key structural features underpin the basis for this activating mutation, thereby providing a molecular rationale for IRF4 K59R -mediated ATLL development.
- John Curtin School of Medical Research, Australian National University, Canberra, Australia.
Organizational Affiliation: