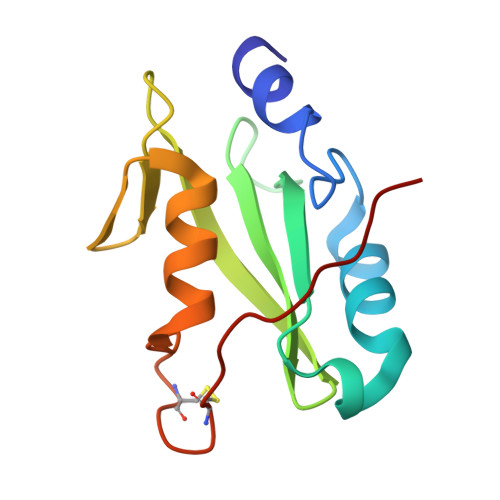
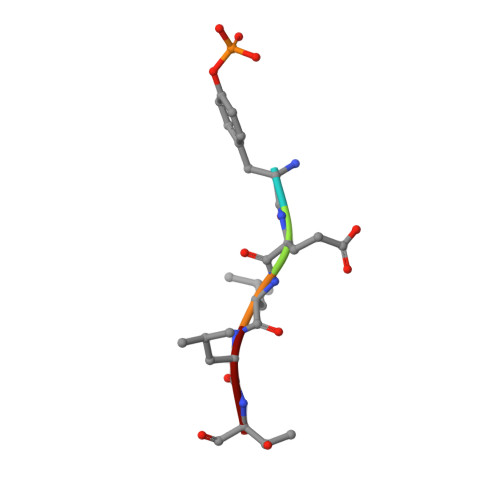
Structural and functional analysis of target recognition by the lymphocyte adaptor protein LNK.
Morris, R., Zhang, Y., Ellyard, J.I., Vinuesa, C.G., Murphy, J.M., Laktyushin, A., Kershaw, N.J., Babon, J.J.(2021) Nat Commun 12: 6110-6110
- PubMed: 34671038
- DOI: https://doi.org/10.1038/s41467-021-26394-6
- Primary Citation of Related Structures:
7R8W, 7R8X - PubMed Abstract:
The SH2B family of adaptor proteins, SH2-B, APS, and LNK are key modulators of cellular signalling pathways. Whilst SH2-B and APS have been partially structurally and biochemically characterised, to date there has been no such characterisation of LNK. Here we present two crystal structures of the LNK substrate recognition domain, the SH2 domain, bound to phosphorylated motifs from JAK2 and EPOR, and biochemically define the basis for target recognition. The LNK SH2 domain adopts a canonical SH2 domain fold with an additional N-terminal helix. Targeted analysis of binding to phosphosites in signalling pathways indicated that specificity is conferred by amino acids one- and three-residues downstream of the phosphotyrosine. Several mutations in LNK showed impaired target binding in vitro and a reduced ability to inhibit signalling, allowing an understanding of the molecular basis of LNK dysfunction in variants identified in patients with myeloproliferative disease.
- Walter and Eliza Hall Institute of Medical Research, 1G Royal Parade, Parkville, VIC, 3052, Australia.
Organizational Affiliation: