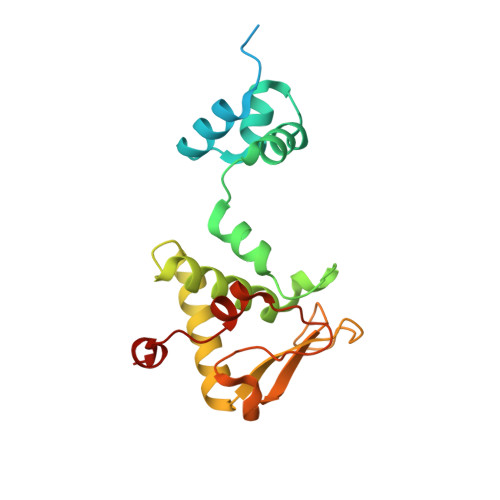
A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
McGinnis, R.J., Brambley, C.A., Stamey, B., Green, W.C., Gragg, K.N., Cafferty, E.R., Terwilliger, T.C., Hammel, M., Hollis, T.J., Miller, J.M., Gainey, M.D., Wallen, J.R.(2022) Nat Commun 13: 4105-4105
- PubMed: 35835745
- DOI: https://doi.org/10.1038/s41467-022-31678-6
- Primary Citation of Related Structures:
7R6R, 7TZ1 - PubMed Abstract:
Regulation of bacteriophage gene expression involves repressor proteins that bind and downregulate early lytic promoters. A large group of mycobacteriophages code for repressors that are unusual in also terminating transcription elongation at numerous binding sites (stoperators) distributed across the phage genome. Here we provide the X-ray crystal structure of a mycobacteriophage immunity repressor bound to DNA, which reveals the binding of a monomer to an asymmetric DNA sequence using two independent DNA binding domains. The structure is supported by small-angle X-ray scattering, DNA binding, molecular dynamics, and in vivo immunity assays. We propose a model for how dual DNA binding domains facilitate regulation of both transcription initiation and elongation, while enabling evolution of other superinfection immune specificities.
- Western Carolina University, Department of Chemistry and Physics, 111 Memorial Drive, Cullowhee, NC, 28723, USA.
Organizational Affiliation: