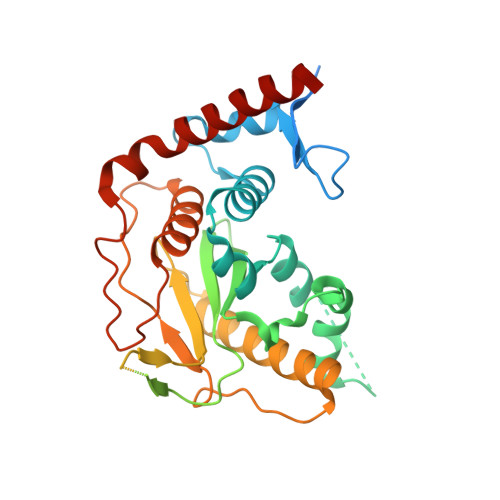
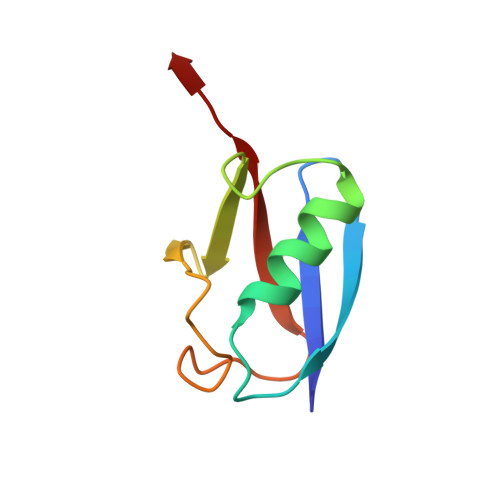
Structural Basis for the SUMO2 Isoform Specificity of SENP7.
Li, Y., De Bolos, A., Amador, V., Reverter, D.(2022) J Mol Biology 434: 167875-167875
- PubMed: 36334780
- DOI: https://doi.org/10.1016/j.jmb.2022.167875
- Primary Citation of Related Structures:
7R2E - PubMed Abstract:
SUMO proteases or deSUMOylases regulate the lifetime of SUMO-conjugated targets in the cell by cleaving off the isopetidic bond between the substrate and the SUMO modifier, thus reversing the conjugation activity of the SUMO E3 ligases. In humans the deSUMOylating activity is mainly conducted by the SENP/ULP protease family, which is constituted of six members sharing a homologous catalytic globular domain. SENP6 and SENP7 are the most divergent members of the family and they show a unique SUMO2/3 isoform preference and a particular activity for dismantling polySUMO2 chains. Here, we present the crystal structure of the catalytic domain of human SENP7 bound to SUMO2, revealing structural key elements for the SUMO2 isoform specificity of SENP7. In particular, we describe the specific contacts between SUMO2 and a unique insertion in SENP7 (named Loop1) that is responsible for the SUMO2 isoform specificity. All the other interface contacts between SENP7 and SUMO2, including the SUMO2 C-terminal tail interaction, are conserved among members of the SENP/ULP family. Our data give insight into an evolutionary adaptation to restrict the deSUMOylating activity in SENP6 and SENP7 for the SUMO2/3 isoforms.
- Institut de Biotecnologia i de Biomedicina (IBB) and Dept. de Bioquímica i Biologia Molecular, Universitat Autònoma de Barcelona, 08193 Bellaterra, Spain.
Organizational Affiliation: