The mRubyFT Protein, Genetically Encoded Blue-to-Red Fluorescent Timer.
Subach, O.M., Tashkeev, A., Vlaskina, A.V., Petrenko, D.E., Gaivoronskii, F.A., Nikolaeva, A.Y., Ivashkina, O.I., Anokhin, K.V., Popov, V.O., Boyko, K.M., Subach, F.V.(2022) Int J Mol Sci 23
- PubMed: 35328628
- DOI: https://doi.org/10.3390/ijms23063208
- Primary Citation of Related Structures:
7QGK - PubMed Abstract:
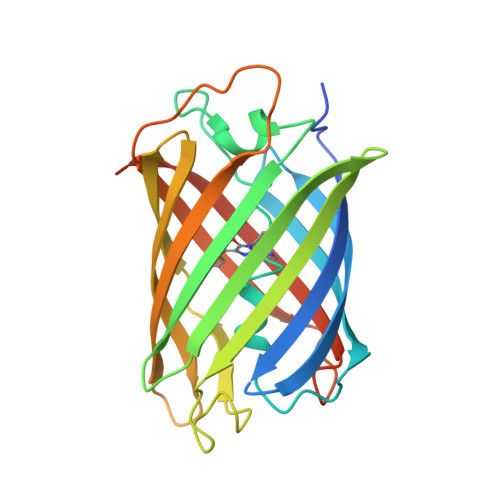
Genetically encoded monomeric blue-to-red fluorescent timers (mFTs) change their fluorescent color over time. mCherry-derived mFTs were used for the tracking of the protein age, visualization of the protein trafficking, and labeling of engram cells. However, the brightness of the blue and red forms of mFTs are 2-3- and 5-7-fold dimmer compared to the brightness of the enhanced green fluorescent protein (EGFP). To address this limitation, we developed a blue-to-red fluorescent timer, named mRubyFT, derived from the bright mRuby2 red fluorescent protein. The blue form of mRubyFT reached its maximum at 5.7 h and completely transformed into the red form that had a maturation half-time of 15 h. Blue and red forms of purified mRubyFT were 4.1-fold brighter and 1.3-fold dimmer than the respective forms of the mCherry-derived Fast-FT timer in vitro. When expressed in mammalian cells, both forms of mRubyFT were 1.3-fold brighter than the respective forms of Fast-FT. The violet light-induced blue-to-red photoconversion was 4.2-fold less efficient in the case of mRubyFT timer compared to the same photoconversion of the Fast-FT timer. The timer behavior of mRubyFT was confirmed in mammalian cells. The monomeric properties of mRubyFT allowed the labeling and confocal imaging of cytoskeleton proteins in live mammalian cells. The X-ray structure of the red form of mRubyFT at 1.5 Å resolution was obtained and analyzed. The role of the residues from the chromophore surrounding was studied using site-directed mutagenesis.
- Complex of NBICS Technologies, National Research Center "Kurchatov Institute", 123182 Moscow, Russia.
Organizational Affiliation: