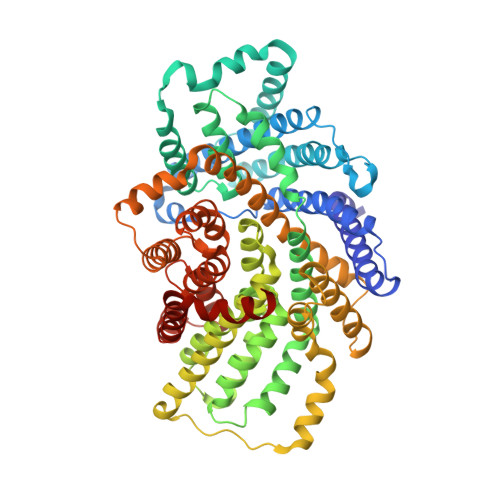
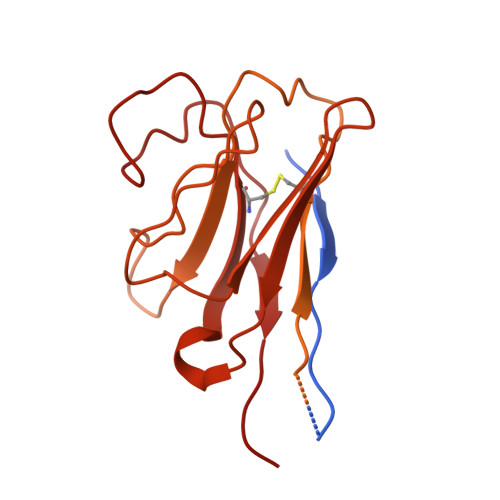
Structural and mechanistic analysis of a tripartite ATP-independent periplasmic TRAP transporter.
Peter, M.F., Ruland, J.A., Depping, P., Schneberger, N., Severi, E., Moecking, J., Gatterdam, K., Tindall, S., Durand, A., Heinz, V., Siebrasse, J.P., Koenig, P.A., Geyer, M., Ziegler, C., Kubitscheck, U., Thomas, G.H., Hagelueken, G.(2022) Nat Commun 13: 4471-4471
- PubMed: 35927235
- DOI: https://doi.org/10.1038/s41467-022-31907-y
- Primary Citation of Related Structures:
7QE5 - PubMed Abstract:
Tripartite ATP-independent periplasmic (TRAP) transporters are found widely in bacteria and archaea and consist of three structural domains, a soluble substrate-binding protein (P-domain), and two transmembrane domains (Q- and M-domains). HiSiaPQM and its homologs are TRAP transporters for sialic acid and are essential for host colonization by pathogenic bacteria. Here, we reconstitute HiSiaQM into lipid nanodiscs and use cryo-EM to reveal the structure of a TRAP transporter. It is composed of 16 transmembrane helices that are unexpectedly structurally related to multimeric elevator-type transporters. The idiosyncratic Q-domain of TRAP transporters enables the formation of a monomeric elevator architecture. A model of the tripartite PQM complex is experimentally validated and reveals the coupling of the substrate-binding protein to the transporter domains. We use single-molecule total internal reflection fluorescence (TIRF) microscopy in solid-supported lipid bilayers and surface plasmon resonance to study the formation of the tripartite complex and to investigate the impact of interface mutants. Furthermore, we characterize high-affinity single variable domains on heavy chain (VHH) antibodies that bind to the periplasmic side of HiSiaQM and inhibit sialic acid uptake, providing insight into how TRAP transporter function might be inhibited in vivo.
- Institute of Structural Biology, University of Bonn, Venusberg-Campus 1, 53127, Bonn, Germany.
Organizational Affiliation: