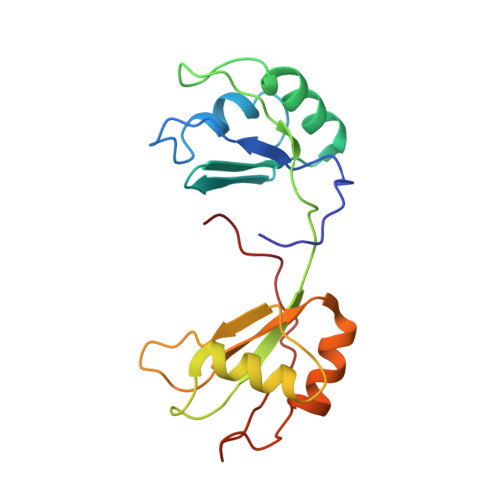
RNA recognition by Npl3p reveals U2 snRNA-binding compatible with a chaperone role during splicing.
Moursy, A., Clery, A., Gerhardy, S., Betz, K.M., Rao, S., Mazur, J., Campagne, S., Beusch, I., Duszczyk, M.M., Robinson, M.D., Panse, V.G., Allain, F.H.(2023) Nat Commun 14: 7166-7166
- PubMed: 37935663
- DOI: https://doi.org/10.1038/s41467-023-42962-4
- Primary Citation of Related Structures:
7QDD, 7QDE - PubMed Abstract:
The conserved SR-like protein Npl3 promotes splicing of diverse pre-mRNAs. However, the RNA sequence(s) recognized by the RNA Recognition Motifs (RRM1 & RRM2) of Npl3 during the splicing reaction remain elusive. Here, we developed a split-iCRAC approach in yeast to uncover the consensus sequence bound to each RRM. High-resolution NMR structures show that RRM2 recognizes a 5´-GNGG-3´ motif leading to an unusual mille-feuille topology. These structures also reveal how RRM1 preferentially interacts with a CC-dinucleotide upstream of this motif, and how the inter-RRM linker and the region C-terminal to RRM2 contribute to cooperative RNA-binding. Structure-guided functional studies show that Npl3 genetically interacts with U2 snRNP specific factors and we provide evidence that Npl3 melts U2 snRNA stem-loop I, a prerequisite for U2/U6 duplex formation within the catalytic center of the B act spliceosomal complex. Thus, our findings suggest an unanticipated RNA chaperoning role for Npl3 during spliceosome active site formation.
- Department of Biology, Institute of Biochemistry, ETH Zurich, Switzerland.
Organizational Affiliation: