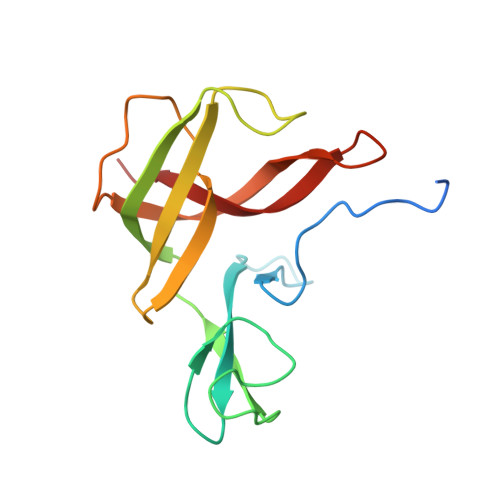
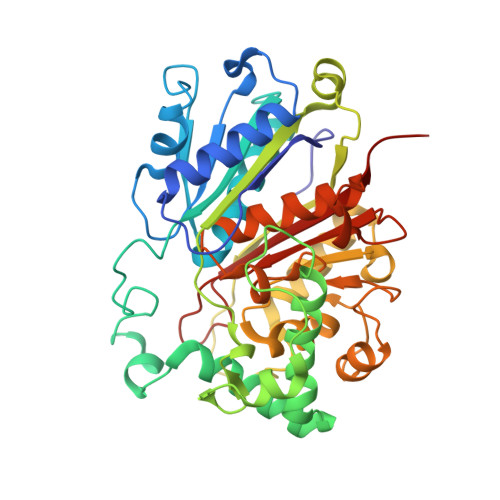
Finis tolueni: a new type of thiolase with an integrated Zn-finger subunit catalyzes the final step of anaerobic toluene metabolism.
Weidenweber, S., Schuhle, K., Lippert, M.L., Mock, J., Seubert, A., Demmer, U., Ermler, U., Heider, J.(2022) FEBS J 289: 5599-5616
- PubMed: 35313080
- DOI: https://doi.org/10.1111/febs.16443
- Primary Citation of Related Structures:
7PXP, 7PYT, 7YXM - PubMed Abstract:
Anaerobic toluene degradation involves β-oxidation of the first intermediate (R)-2-benzylsuccinate to succinyl-CoA and benzoyl-CoA. Here, we characterize the last enzyme of this pathway, (S)-2-benzoylsuccinyl-CoA thiolase (BbsAB). Although benzoylsuccinyl-CoA is not available for enzyme assays, the recombinantly produced enzymes from two different species showed the reverse activity, benzoylsuccinyl-CoA formation from benzoyl-CoA and succinyl-CoA. Activity depended on the presence of both subunits, the thiolase family member BbsB and the Zn-finger protein BbsA, which is affiliated to the DUF35 family of unknown function. We determined the structure of BbsAB from Geobacter metallireducens with and without bound CoA at 1.7 and 2.0 Å resolution, respectively. CoA binding into the well-known thiolase cavity triggers an induced-fit movement of the highly disordered covering loop, resulting in its rigidification by forming multiple interactions to the outstretched CoA moiety. This event is coupled with an 8 Å movement of an adjacent hairpin loop of BbsB and the C-terminal domain of BbsA. Thereby, CoA is placed into a catalytically productive conformation, and a putative second CoA binding site involving BbsA and the partner BbsB' subunit is simultaneously formed that also reaches the active center. Therefore, while maintaining the standard thioester-dependent Claisen-type mechanism, BbsAB represents a new type of thiolase.
- Max-Planck-Institut für Biophysik, Frankfurt am Main, Germany.
Organizational Affiliation: