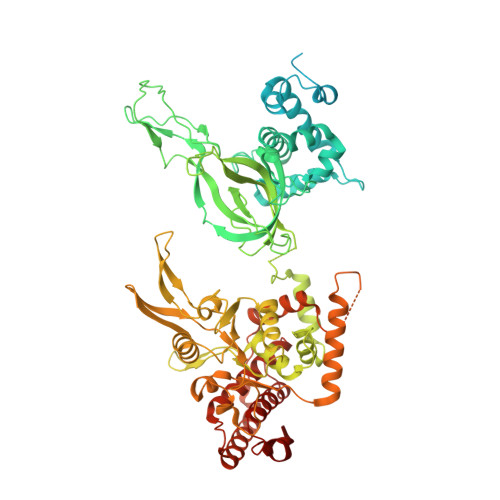
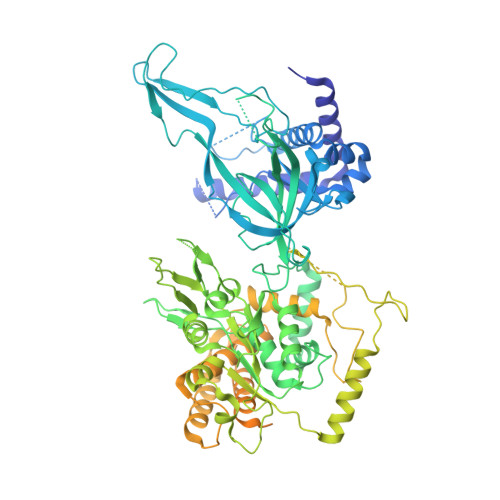
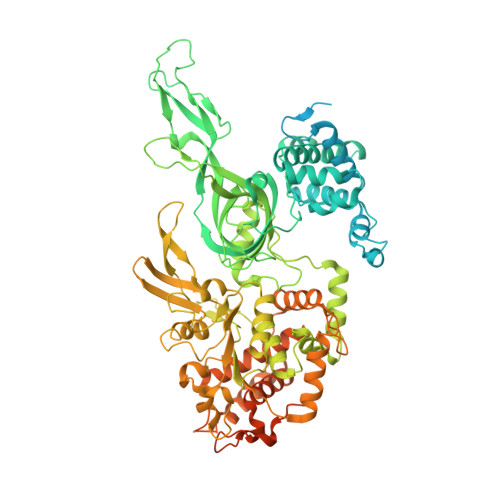
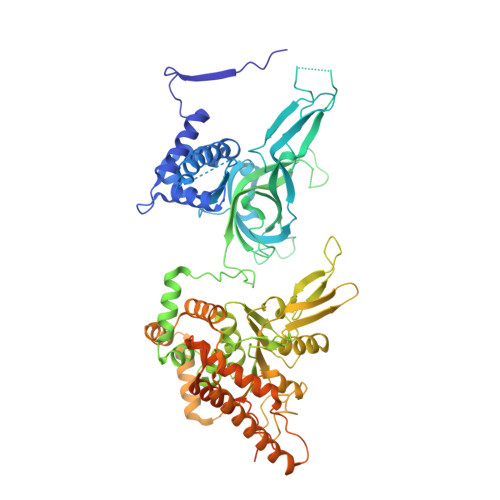
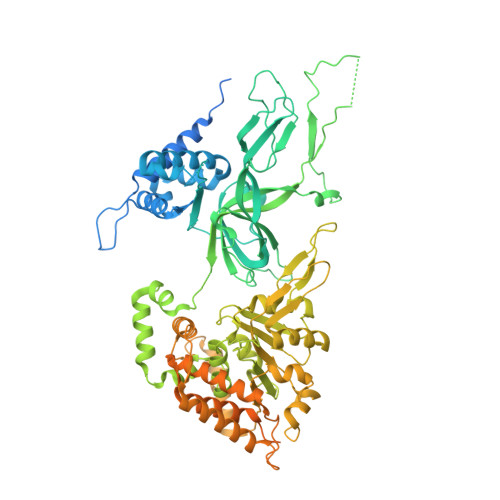
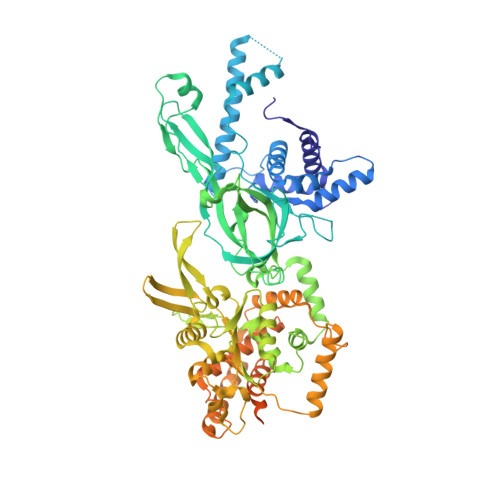
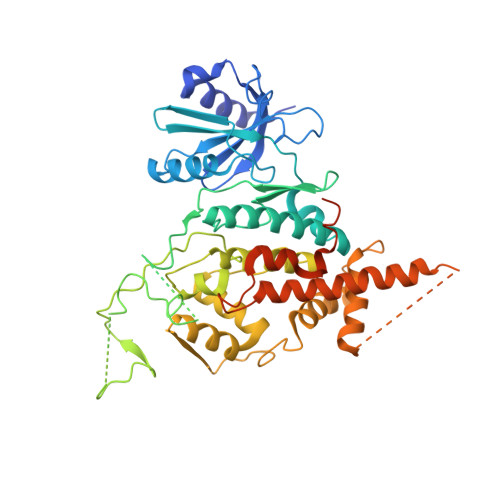
The structural basis of Cdc7-Dbf4 kinase dependent targeting and phosphorylation of the MCM2-7 double hexamer.
Saleh, A., Noguchi, Y., Aramayo, R., Ivanova, M.E., Stevens, K.M., Montoya, A., Sunidhi, S., Carranza, N.L., Skwark, M.J., Speck, C.(2022) Nat Commun 13: 2915-2915
- PubMed: 35614055
- DOI: https://doi.org/10.1038/s41467-022-30576-1
- Primary Citation of Related Structures:
7PT6, 7PT7 - PubMed Abstract:
The controlled assembly of replication forks is critical for genome stability. The Dbf4-dependent Cdc7 kinase (DDK) initiates replisome assembly by phosphorylating the MCM2-7 replicative helicase at the N-terminal tails of Mcm2, Mcm4 and Mcm6. At present, it remains poorly understood how DDK docks onto the helicase and how the kinase targets distal Mcm subunits for phosphorylation. Using cryo-electron microscopy and biochemical analysis we discovered that an interaction between the HBRCT domain of Dbf4 with Mcm2 serves as an anchoring point, which supports binding of DDK across the MCM2-7 double-hexamer interface and phosphorylation of Mcm4 on the opposite hexamer. Moreover, a rotation of DDK along its anchoring point allows phosphorylation of Mcm2 and Mcm6. In summary, our work provides fundamental insights into DDK structure, control and selective activation of the MCM2-7 helicase during DNA replication. Importantly, these insights can be exploited for development of novel DDK inhibitors.
- DNA Replication Group, Institute of Clinical Sciences, Faculty of Medicine, Imperial College London, Du Cane Road, London, W12 0NN, UK.
Organizational Affiliation: