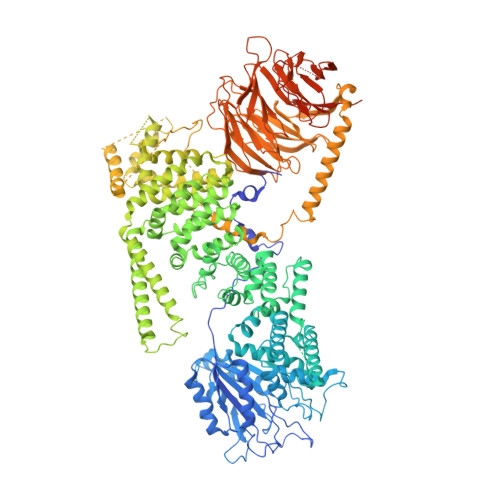
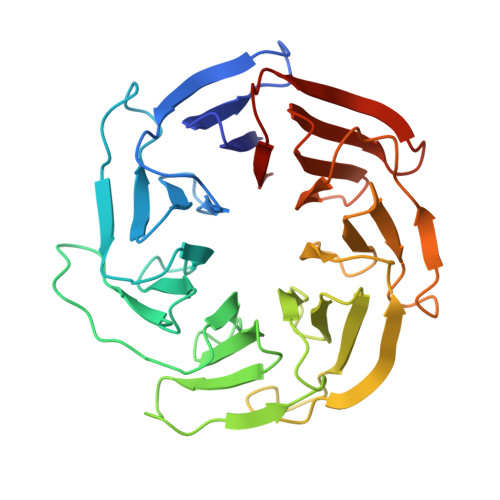
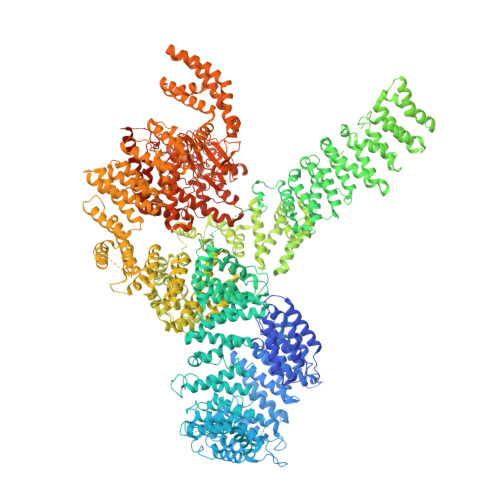
EGOC inhibits TOROID polymerization by structurally activating TORC1.
Prouteau, M., Bourgoint, C., Felix, J., Bonadei, L., Sadian, Y., Gabus, C., Savvides, S.N., Gutsche, I., Desfosses, A., Loewith, R.(2023) Nat Struct Mol Biol 30: 273-285
- PubMed: 36702972
- DOI: https://doi.org/10.1038/s41594-022-00912-6
- Primary Citation of Related Structures:
7PQH - PubMed Abstract:
Target of rapamycin complex 1 (TORC1) is a protein kinase controlling cell homeostasis and growth in response to nutrients and stresses. In Saccharomyces cerevisiae, glucose depletion triggers a redistribution of TORC1 from a dispersed localization over the vacuole surface into a large, inactive condensate called TOROID (TORC1 organized in inhibited domains). However, the mechanisms governing this transition have been unclear. Here, we show that acute depletion and repletion of EGO complex (EGOC) activity is sufficient to control TOROID distribution, independently of other nutrient-signaling pathways. The 3.9-Å-resolution structure of TORC1 from TOROID cryo-EM data together with interrogation of key interactions in vivo provide structural insights into TORC1-TORC1' and TORC1-EGOC interaction interfaces. These data support a model in which glucose-dependent activation of EGOC triggers binding to TORC1 at an interface required for TOROID assembly, preventing TORC1 polymerization and promoting release of active TORC1.
- Department of Molecular and Cellular Biology, University of Geneva, Geneva, Switzerland. manoel.prouteau@unige.ch.
Organizational Affiliation: