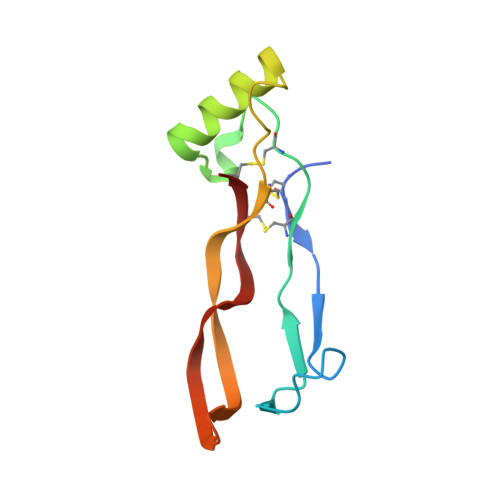
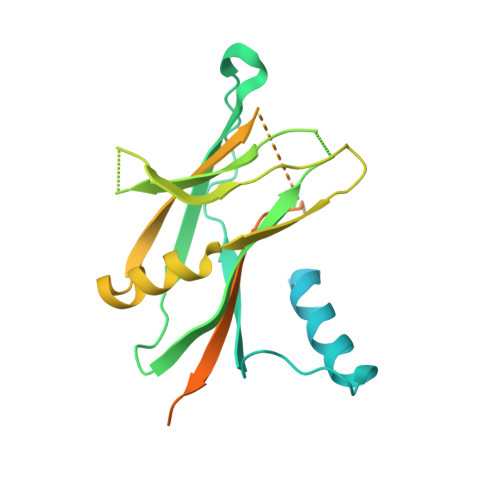
Crystal structures of BMPRII extracellular domain in binary and ternary receptor complexes with BMP10.
Guo, J., Liu, B., Thorikay, M., Yu, M., Li, X., Tong, Z., Salmon, R.M., Read, R.J., Ten Dijke, P., Morrell, N.W., Li, W.(2022) Nat Commun 13: 2395-2395
- PubMed: 35504921
- DOI: https://doi.org/10.1038/s41467-022-30111-2
- Primary Citation of Related Structures:
7POI, 7POJ, 7PPA, 7PPB, 7PPC - PubMed Abstract:
Heterozygous mutations in BMPR2 (bone morphogenetic protein (BMP) receptor type II) cause pulmonary arterial hypertension. BMPRII is a receptor for over 15 BMP ligands, but why BMPR2 mutations cause lung-specific pathology is unknown. To elucidate the molecular basis of BMP:BMPRII interactions, we report crystal structures of binary and ternary BMPRII receptor complexes with BMP10, which contain an ensemble of seven different BMP10:BMPRII 1:1 complexes. BMPRII binds BMP10 at the knuckle epitope, with the A-loop and β4 strand making BMPRII-specific interactions. The BMPRII binding surface on BMP10 is dynamic, and the affinity is weaker in the ternary complex than in the binary complex. Hydrophobic core and A-loop interactions are important in BMPRII-mediated signalling. Our data reveal how BMPRII is a low affinity receptor, implying that forming a signalling complex requires high concentrations of BMPRII, hence mutations will impact on tissues with highest BMPR2 expression such as the lung vasculature.
- Department of Medicine, University of Cambridge School of Clinical Medicine, Cambridge, CB2 0QQ, United Kingdom.
Organizational Affiliation: