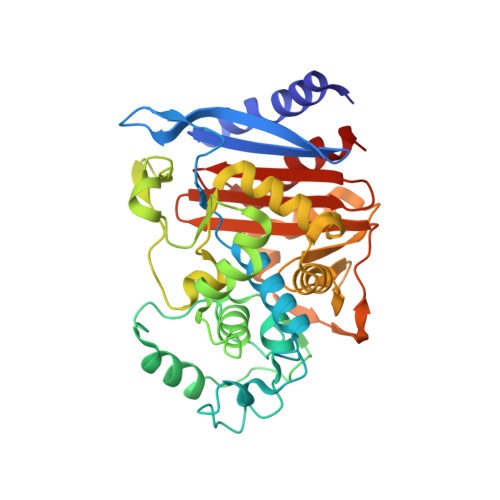
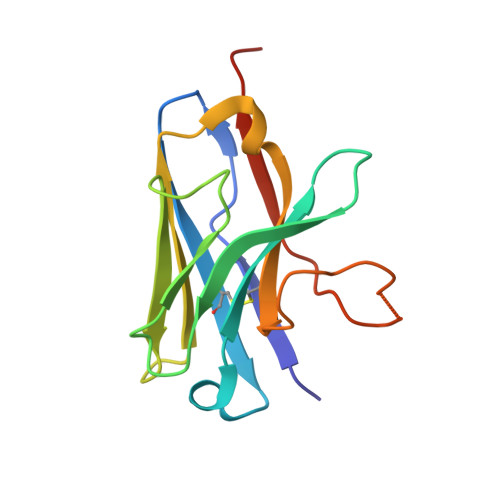
Development of Nanobodies as Theranostic Agents against CMY-2-Like Class C beta-Lactamases.
Cawez, F., Mercuri, P.S., Morales-Yanez, F.J., Maalouf, R., Vandevenne, M., Kerff, F., Guerin, V., Mainil, J., Thiry, D., Saulmont, M., Vanderplasschen, A., Lafaye, P., Ayme, G., Bogaerts, P., Dumoulin, M., Galleni, M.(2023) Antimicrob Agents Chemother 67: e0149922-e0149922
- PubMed: 36892280
- DOI: https://doi.org/10.1128/aac.01499-22
- Primary Citation of Related Structures:
7PA5 - PubMed Abstract:
Three soluble single-domain fragments derived from the unique variable region of camelid heavy-chain antibodies (VHHs) against the CMY-2 β-lactamase behaved as inhibitors. The structure of the complex VHH cAb CMY-2 (254)/CMY-2 showed that the epitope is close to the active site and that the CDR3 of the VHH protrudes into the catalytic site. The β-lactamase inhibition pattern followed a mixed profile with a predominant noncompetitive component. The three isolated VHHs recognized overlapping epitopes since they behaved as competitive binders. Our study identified a binding site that can be targeted by a new class of β-lactamase inhibitors designed on the sequence of the paratope. Furthermore, the use of mono- or bivalent VHH and rabbit polyclonal anti-CMY-2 antibodies enables the development of the first generation of enzyme-linked immunosorbent assay (ELISA) for the detection of CMY-2 produced by CMY-2-expressing bacteria, irrespective of resistotype.
- InBioS, Center for Protein Engineering, Biological Macromolecules, Department of Life Sciences, University of Liège, Liège, Belgium.
Organizational Affiliation: