The phage defence island of a multidrug resistant plasmid uses both BREX and type IV restriction for complementary protection from viruses.
Picton, D.M., Luyten, Y.A., Morgan, R.D., Nelson, A., Smith, D.L., Dryden, D.T.F., Hinton, J.C.D., Blower, T.R.(2021) Nucleic Acids Res 49: 11257-11273
- PubMed: 34657954
- DOI: https://doi.org/10.1093/nar/gkab906
- Primary Citation of Related Structures:
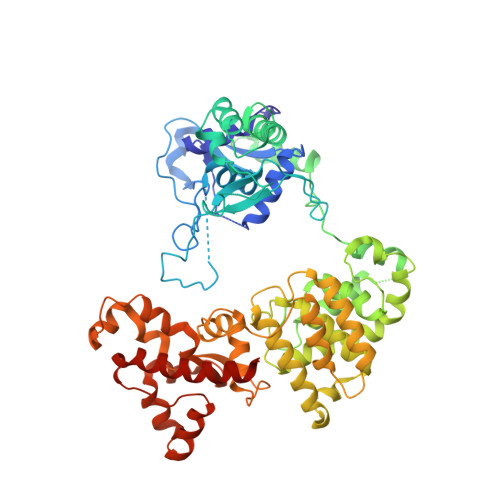
7P9K, 7P9M - PubMed Abstract:
Bacteria have evolved a multitude of systems to prevent invasion by bacteriophages and other mobile genetic elements. Comparative genomics suggests that genes encoding bacterial defence mechanisms are often clustered in 'defence islands', providing a concerted level of protection against a wider range of attackers. However, there is a comparative paucity of information on functional interplay between multiple defence systems. Here, we have functionally characterised a defence island from a multidrug resistant plasmid of the emerging pathogen Escherichia fergusonii. Using a suite of thirty environmentally-isolated coliphages, we demonstrate multi-layered and robust phage protection provided by a plasmid-encoded defence island that expresses both a type I BREX system and the novel GmrSD-family type IV DNA modification-dependent restriction enzyme, BrxU. We present the structure of BrxU to 2.12 Å, the first structure of the GmrSD family of enzymes, and show that BrxU can utilise all common nucleotides and a wide selection of metals to cleave a range of modified DNAs. Additionally, BrxU undergoes a multi-step reaction cycle instigated by an unexpected ATP-dependent shift from an intertwined dimer to monomers. This direct evidence that bacterial defence islands can mediate complementary layers of phage protection enhances our understanding of the ever-expanding nature of phage-bacterial interactions.
- Department of Biosciences, Durham University, Stockton Road, Durham DH1 3LE, UK.
Organizational Affiliation: