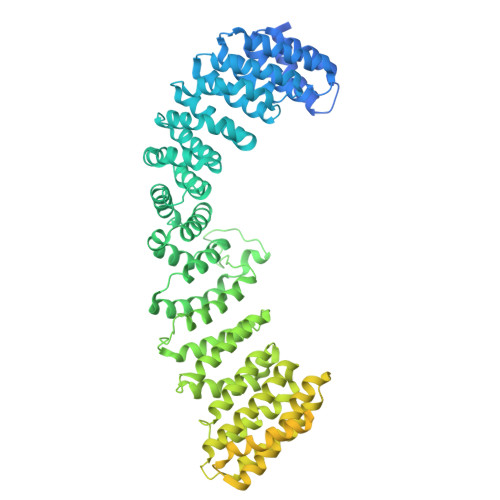
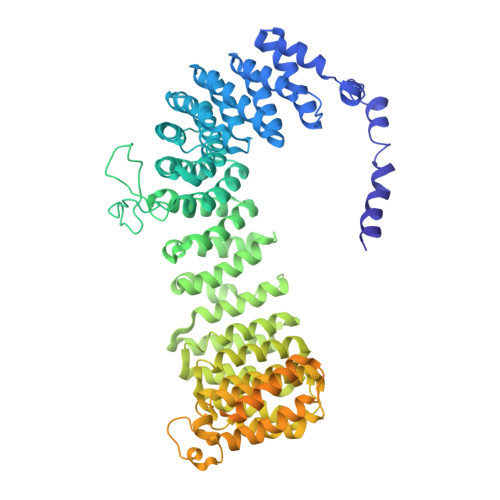
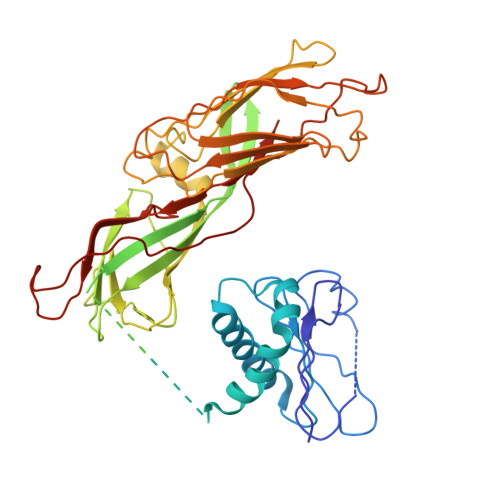
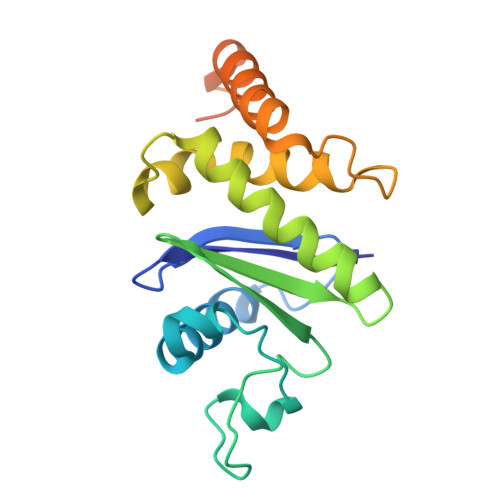
Flexible open conformation of the AP-3 complex explains its role in cargo recruitment at the Golgi.
Schoppe, J., Schubert, E., Apelbaum, A., Yavavli, E., Birkholz, O., Stephanowitz, H., Han, Y., Perz, A., Hofnagel, O., Liu, F., Piehler, J., Raunser, S., Ungermann, C.(2021) J Biological Chem 297: 101334-101334
- PubMed: 34688652
- DOI: https://doi.org/10.1016/j.jbc.2021.101334
- Primary Citation of Related Structures:
7P3X, 7P3Y, 7P3Z - PubMed Abstract:
Vesicle formation at endomembranes requires the selective concentration of cargo by coat proteins. Conserved adapter protein complexes at the Golgi (AP-3), the endosome (AP-1), or the plasma membrane (AP-2) with their conserved core domain and flexible ear domains mediate this function. These complexes also rely on the small GTPase Arf1 and/or specific phosphoinositides for membrane binding. The structural details that influence these processes, however, are still poorly understood. Here we present cryo-EM structures of the full-length stable 300 kDa yeast AP-3 complex. The structures reveal that AP-3 adopts an open conformation in solution, comparable to the membrane-bound conformations of AP-1 or AP-2. This open conformation appears to be far more flexible than AP-1 or AP-2, resulting in compact, intermediate, and stretched subconformations. Mass spectrometrical analysis of the cross-linked AP-3 complex further indicates that the ear domains are flexibly attached to the surface of the complex. Using biochemical reconstitution assays, we also show that efficient AP-3 recruitment to the membrane depends primarily on cargo binding. Once bound to cargo, AP-3 clustered and immobilized cargo molecules, as revealed by single-molecule imaging on polymer-supported membranes. We conclude that its flexible open state may enable AP-3 to bind and collect cargo at the Golgi and could thus allow coordinated vesicle formation at the trans-Golgi upon Arf1 activation.
- Department of Biology/Chemistry, Biochemistry Section, Osnabrück University, Osnabrück, Germany.
Organizational Affiliation: