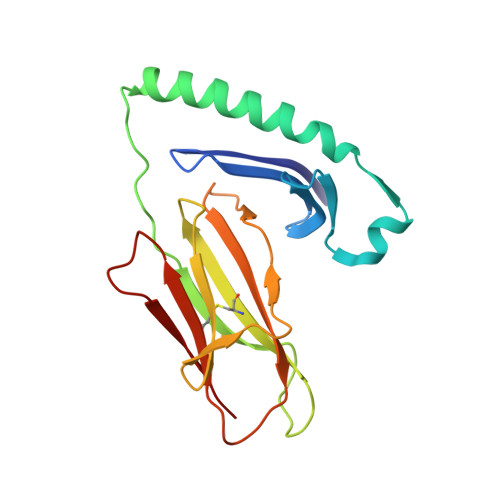
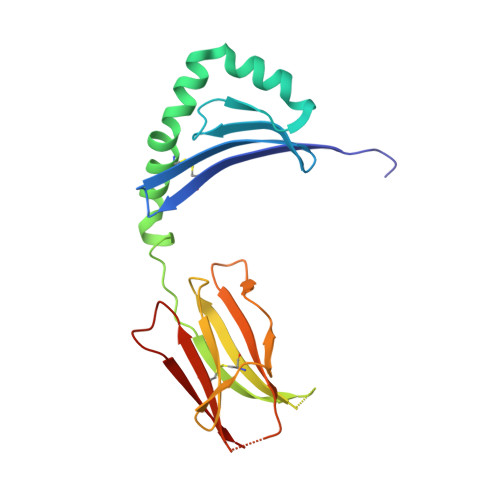
Key interactions in the trimolecular complex consisting of the rheumatoid arthritis-associated DRB1*04:01 molecule, the major glycosylated collagen II peptide and the T-cell receptor.
Ge, C., Weisse, S., Xu, B., Dobritzsch, D., Viljanen, J., Kihlberg, J., Do, N.N., Schneider, N., Lanig, H., Holmdahl, R., Burkhardt, H.(2022) Ann Rheum Dis 81: 480-489
- PubMed: 35027402
- DOI: https://doi.org/10.1136/annrheumdis-2021-220500
- Primary Citation of Related Structures:
7NZE, 7NZF, 7NZH, 7O00 - PubMed Abstract:
Rheumatoid arthritis (RA) is an autoimmune disease strongly associated with the major histocompatibility complex (MHC) class II allele DRB1*04:01, which encodes a protein that binds self-peptides for presentation to T cells. This study characterises the autoantigen-presenting function of DRB1*04:01 (HLA-DRA*01:01/HLA-DRB1*04:01) at a molecular level for prototypic T-cell determinants, focusing on a post-translationally modified collagen type II (Col2)-derived peptide. The crystal structures of DRB1*04:01 molecules in complex with the peptides HSP70 289-306 , citrullinated CILP 982-996 and galactosylated Col2 259-273 were determined on cocrystallisation. T cells specific for Col2 259-273 were investigated in peripheral blood mononuclear cells from patients with DRB1*04:01-positive RA by cytofluorometric detection of the activation marker CD154 on peptide stimulation and binding of fluorescent DRB1*0401/Col2 259-273 tetramer complexes. The cDNAs encoding the T-cell receptor (TCR) α-chains and β-chains were cloned from single-cell sorted tetramer-positive T cells and transferred via a lentiviral vector into TCR-deficient Jurkat 76 cells. The crystal structures identified peptide binding to DRB1*04:01 and potential side chain exposure to T cells. The main TCR recognition sites in Col2 259-273 were lysine residues that can be galactosylated. RA T-cell responses to DRB1*04:01-presented Col2 259-273 were dependent on peptide galactosylation at lysine 264. Dynamic molecular modelling of a functionally characterised Col2 259-273 -specific TCR complexed with DRB1*04:01/Col2 259-273 provided evidence for differential allosteric T-cell recognition of glycosylated lysine 264. The MHC-peptide-TCR interactions elucidated in our study provide new molecular insights into recognition of a post-translationally modified RA T-cell determinant with a known dominant role in arthritogenic and tolerogenic responses in murine Col2-induced arthritis.
- Section for Medical Inflammation Research, Department of Medical Biochemistry and Biophysics, Karolinska Institute, Stockholm, Sweden.
Organizational Affiliation: