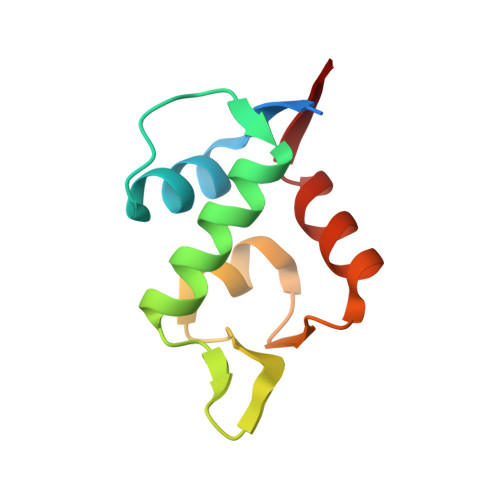
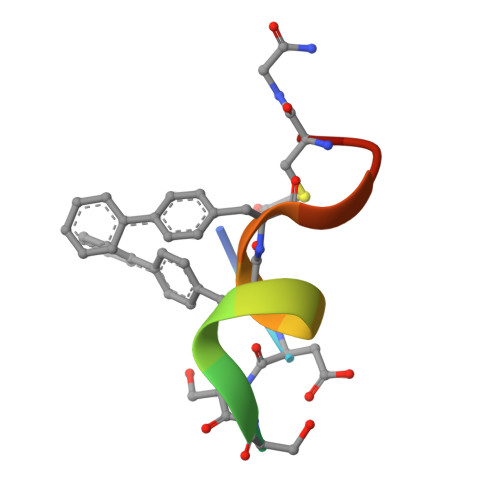
Discovery, X-ray structure and CPP-conjugation enabled uptake of p53/MDM2 macrocyclic peptide inhibitors.
Schneider, A.F.L., Kallen, J., Ottl, J., Reid, P.C., Ripoche, S., Ruetz, S., Stachyra, T.M., Hintermann, S., Dumelin, C.E., Hackenberger, C.P.R., Marzinzik, A.L.(2021) RSC Chem Biol 2: 1661-1668
- PubMed: 34977581
- DOI: https://doi.org/10.1039/d1cb00056j
- Primary Citation of Related Structures:
7NUS - PubMed Abstract:
Mouse double minute 2 homolog (MDM2, Hdm2) is an important negative regulator of the tumor suppressor p53. Using a mRNA based display technique to screen a library of >10 12 in vitro -translated cyclic peptides, we have identified a macrocyclic ligand that shows picomolar potency on MDM2. X-Ray crystallography reveals a novel binding mode utilizing a unique pharmacophore to occupy the Phe/Trp/Leu pockets on MDM2. Conjugation of a cyclic cell-penetrating peptide (cCPP) to the initially non cell-permeable ligand enables cellular uptake and a pharmacodynamic response in SJSA-1 cells. The demonstrated enhanced intracellular availability of cyclic peptides that are identified by a display technology exemplifies a process for the application of intracellular tools for drug discovery projects.
- Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Robert-Rössle-Strasse 10 Berlin 13125 Germany hackenbe@fmp-berlin.de.
Organizational Affiliation: