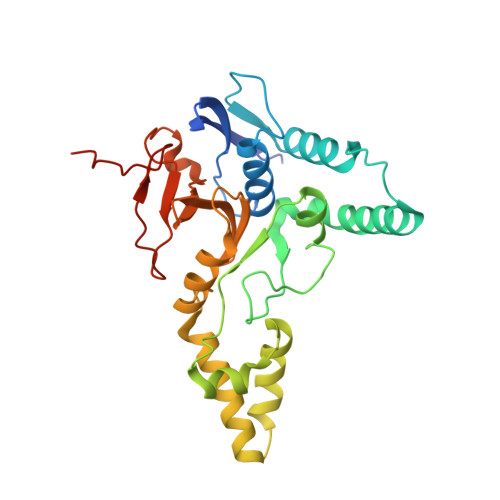
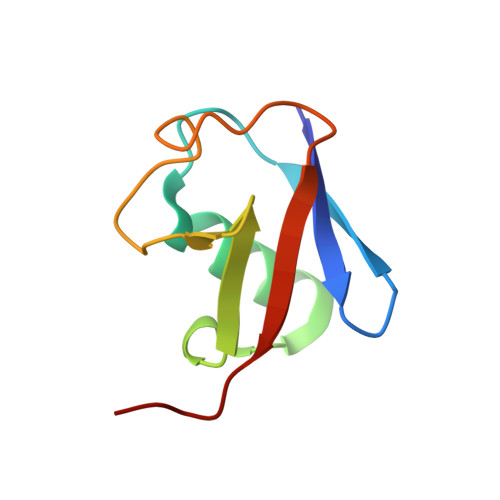
Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Abdul Rehman, S.A., Armstrong, L.A., Lange, S.M., Kristariyanto, Y.A., Grawert, T.W., Knebel, A., Svergun, D.I., Kulathu, Y.(2021) Mol Cell 81: 4176-4190.e6
- PubMed: 34529927
- DOI: https://doi.org/10.1016/j.molcel.2021.08.024
- Primary Citation of Related Structures:
6TUV, 6TXB, 6Y6R, 6YJG, 6Z49, 6Z7V, 6Z90, 7NPI - PubMed Abstract:
Of the eight distinct polyubiquitin (polyUb) linkages that can be assembled, the roles of K48-linked polyUb (K48-polyUb) are the most established, with K48-polyUb modified proteins being targeted for degradation. MINDY1 and MINDY2 are members of the MINDY family of deubiquitinases (DUBs) that have exquisite specificity for cleaving K48-polyUb, yet we have a poor understanding of their catalytic mechanism. Here, we analyze the crystal structures of MINDY1 and MINDY2 alone and in complex with monoUb, di-, and penta-K48-polyUb, identifying 5 distinct Ub binding sites in the catalytic domain that explain how these DUBs sense both Ub chain length and linkage type to cleave K48-polyUb chains. The activity of MINDY1/2 is inhibited by the Cys-loop, and we find that substrate interaction relieves autoinhibition to activate these DUBs. We also find that MINDY1/2 use a non-canonical catalytic triad composed of Cys-His-Thr. Our findings highlight multiple layers of regulation modulating DUB activity in MINDY1 and MINDY2.
- MRC Protein Phosphorylation & Ubiquitylation Unit, School of Life Sciences, University of Dundee, Dow Street, Dundee DD1 5EH, UK. Electronic address: s.z.rehman@dundee.ac.uk.
Organizational Affiliation: