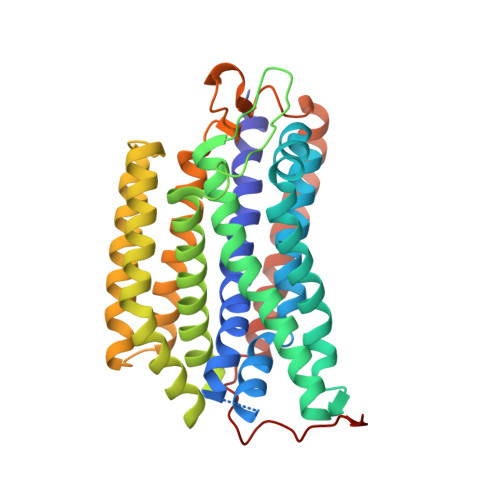
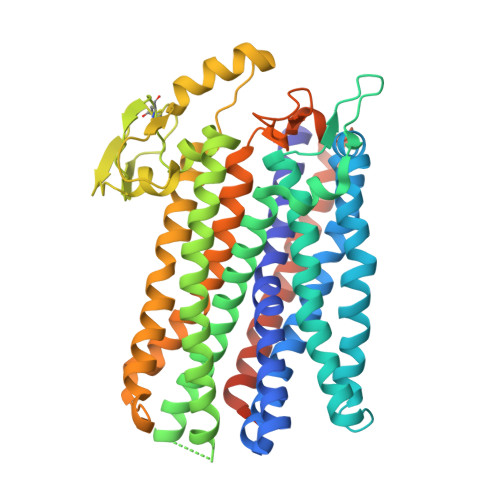
The cryo-EM structure of the bd oxidase from M. tuberculosis reveals a unique structural framework and enables rational drug design to combat TB.
Safarian, S., Opel-Reading, H.K., Wu, D., Mehdipour, A.R., Hards, K., Harold, L.K., Radloff, M., Stewart, I., Welsch, S., Hummer, G., Cook, G.M., Krause, K.L., Michel, H.(2021) Nat Commun 12: 5236-5236
- PubMed: 34475399
- DOI: https://doi.org/10.1038/s41467-021-25537-z
- Primary Citation of Related Structures:
7NKZ - PubMed Abstract:
New drugs are urgently needed to combat the global TB epidemic. Targeting simultaneously multiple respiratory enzyme complexes of Mycobacterium tuberculosis is regarded as one of the most effective treatment options to shorten drug administration regimes, and reduce the opportunity for the emergence of drug resistance. During infection and proliferation, the cytochrome bd oxidase plays a crucial role for mycobacterial pathophysiology by maintaining aerobic respiration at limited oxygen concentrations. Here, we present the cryo-EM structure of the cytochrome bd oxidase from M. tuberculosis at 2.5 Å. In conjunction with atomistic molecular dynamics (MD) simulation studies we discovered a previously unknown MK-9-binding site, as well as a unique disulfide bond within the Q-loop domain that defines an inactive conformation of the canonical quinol oxidation site in Actinobacteria. Our detailed insights into the long-sought atomic framework of the cytochrome bd oxidase from M. tuberculosis will form the basis for the design of highly specific drugs to act on this enzyme.
- Department of Molecular Membrane Biology, Max Planck Institute of Biophysics, Frankfurt/Main, Germany. schara.safarian@biophys.mpg.de.
Organizational Affiliation: