OC43 coronavirus methyltransferase
Dostalik, P., Krafcikova, P., Boura, E.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Replicase polyprotein 1ab | 299 | Human coronavirus OC43 | Mutation(s): 0 Gene Names: rep, 1a-1b EC: 3.4.19.12 (PDB Primary Data), 3.4.22.69 (PDB Primary Data), 3.4.22 (PDB Primary Data), 2.7.7.48 (PDB Primary Data), 3.6.4.12 (PDB Primary Data), 3.6.4.13 (PDB Primary Data), 2.1.1 (PDB Primary Data), 3.1.13 (PDB Primary Data), 3.1 (PDB Primary Data) | 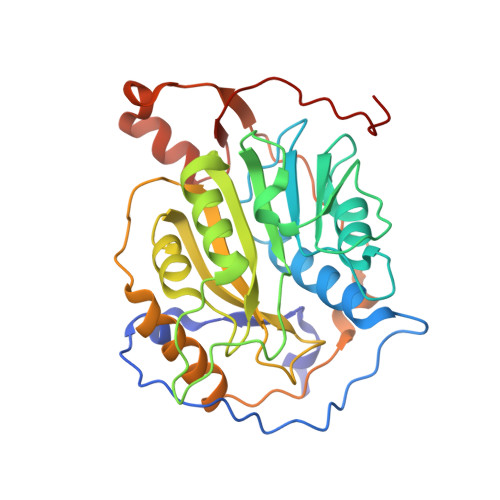 | |
UniProt | |||||
Find proteins for P0C6X6 (Human coronavirus OC43) Explore P0C6X6 Go to UniProtKB: P0C6X6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C6X6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Replicase polyprotein 1a | 122 | Human coronavirus OC43 | Mutation(s): 0 Gene Names: 1a EC: 3.4.19.12 (PDB Primary Data), 3.4.22.69 (PDB Primary Data), 3.4.22 (PDB Primary Data) | 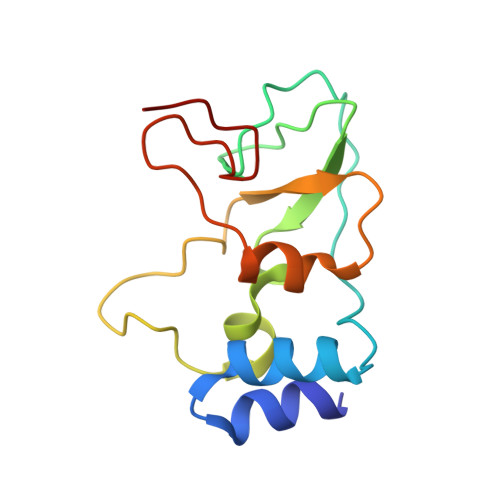 | |
UniProt | |||||
Find proteins for P0C6U7 (Human coronavirus OC43) Explore P0C6U7 Go to UniProtKB: P0C6U7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C6U7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SFG Query on SFG | C [auth A] | SINEFUNGIN C15 H23 N7 O5 LMXOHSDXUQEUSF-YECHIGJVSA-N |  | ||
| ZN Query on ZN | D [auth B], E [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 61.945 | α = 90 |
| b = 61.945 | β = 90 |
| c = 109.717 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PHENIX | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |