Design of proteasome inhibitors with oral efficacy in vivo against Plasmodium falciparum and selectivity over the human proteasome.
Xie, S.C., Metcalfe, R.D., Mizutani, H., Puhalovich, T., Hanssen, E., Morton, C.J., Du, Y., Dogovski, C., Huang, S.C., Ciavarri, J., Hales, P., Griffin, R.J., Cohen, L.H., Chuang, B.C., Wittlin, S., Deni, I., Yeo, T., Ward, K.E., Barry, D.C., Liu, B., Gillett, D.L., Crespo-Fernandez, B.F., Ottilie, S., Mittal, N., Churchyard, A., Ferguson, D., Aguiar, A.C.C., Guido, R.V.C., Baum, J., Hanson, K.K., Winzeler, E.A., Gamo, F.J., Fidock, D.A., Baud, D., Parker, M.W., Brand, S., Dick, L.R., Griffin, M.D.W., Gould, A.E., Tilley, L.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34548400
- DOI: https://doi.org/10.1073/pnas.2107213118
- Primary Citation of Related Structures:
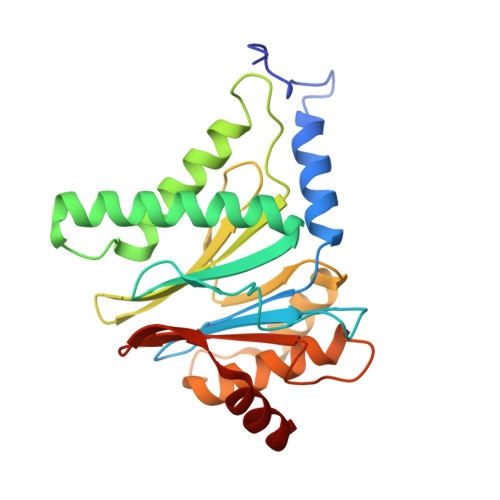
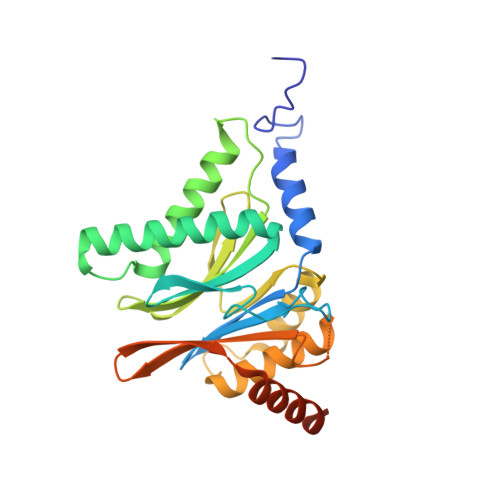
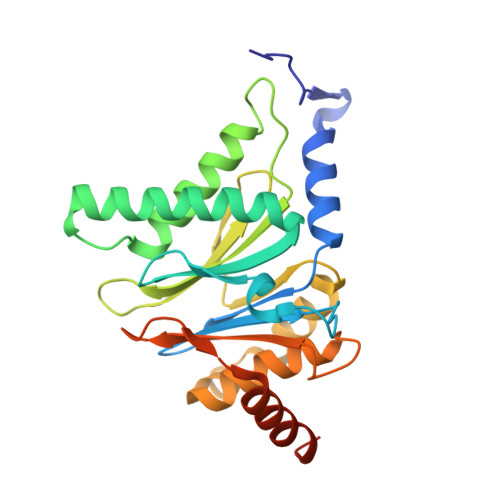
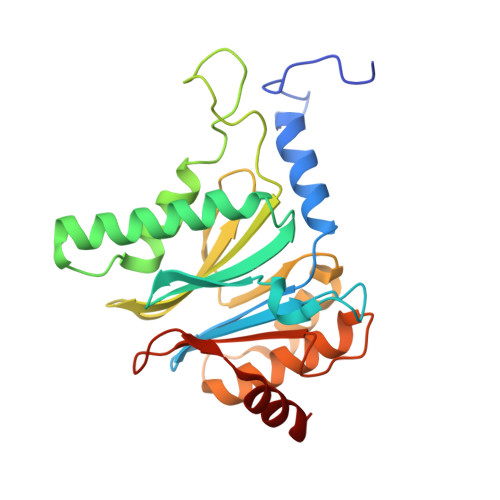
7LXT, 7LXU, 7LXV - PubMed Abstract:
The Plasmodium falciparum proteasome is a potential antimalarial drug target. We have identified a series of amino-amide boronates that are potent and specific inhibitors of the P. falciparum 20S proteasome ( Pf 20S) β5 active site and that exhibit fast-acting antimalarial activity. They selectively inhibit the growth of P. falciparum compared with a human cell line and exhibit high potency against field isolates of P. falciparum and Plasmodium vivax They have a low propensity for development of resistance and possess liver stage and transmission-blocking activity. Exemplar compounds, MPI-5 and MPI-13, show potent activity against P. falciparum infections in a SCID mouse model with an oral dosing regimen that is well tolerated. We show that MPI-5 binds more strongly to Pf 20S than to human constitutive 20S ( Hs 20Sc). Comparison of the cryo-electron microscopy (EM) structures of Pf 20S and Hs 20Sc in complex with MPI-5 and Pf 20S in complex with the clinically used anti-cancer agent, bortezomib, reveal differences in binding modes that help to explain the selectivity. Together, this work provides insights into the 20S proteasome in P. falciparum , underpinning the design of potent and selective antimalarial proteasome inhibitors.
- Department of Biochemistry and Pharmacology, Bio21 Molecular Science and Biotechnology Institute, The University of Melbourne, Melbourne, VIC 3010, Australia.
Organizational Affiliation: