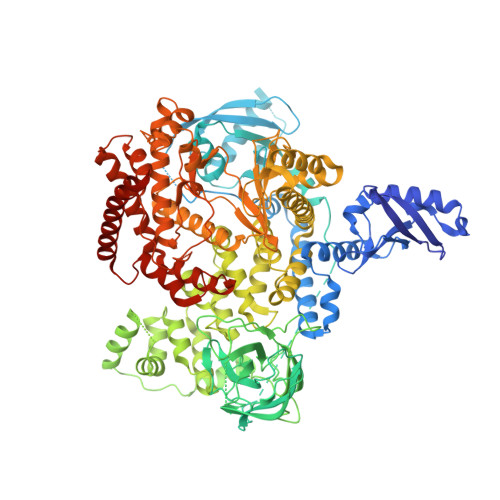
Discovery of a new series of PI3K-delta inhibitors from Virtual Screening.
Fradera, X., Deng, Q., Achab, A., Garcia, Y., Kattar, S.D., McGowan, M.A., Methot, J.L., Wilson, K., Zhou, H., Shaffer, L., Goldenblatt, P., Tong, V., Augustin, M.A., Altman, M.D., Lesburg, C.A., Shah, S., Katz, J.D.(2021) Bioorg Med Chem Lett 42: 128046-128046
- PubMed: 33865969
- DOI: https://doi.org/10.1016/j.bmcl.2021.128046
- Primary Citation of Related Structures:
7LQ1 - PubMed Abstract:
PI3K-δ mediates key immune cell signaling pathways and is a target of interest for treatment of oncological and immunological disorders. Here we describe the discovery and optimization of a novel series of PI3K-δ selective inhibitors. We first identified hits containing an isoindolinone scaffold using a combined ligand- and receptor-based virtual screening workflow, and then improved potency and selectivity guided by structural data and modeling. Careful optimization of molecular properties led to compounds with improved permeability and pharmacokinetic profile, and high potency in a whole blood assay.
- Computational and Structural Chemistry, Merck & Co., Inc., Boston, MA, USA. Electronic address: xavier.fradera@merck.com.
Organizational Affiliation: