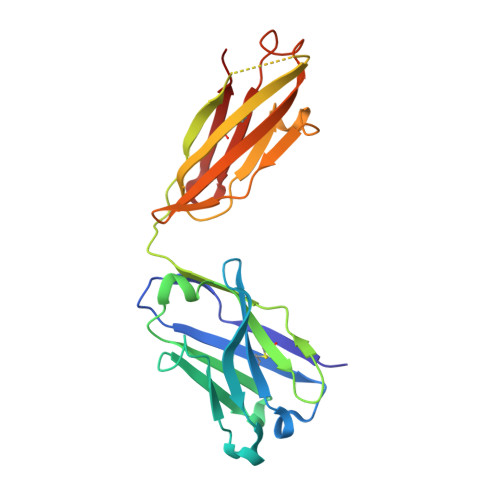
A conserved epitope III on hepatitis C virus E2 protein has alternate conformations facilitating cell binding or virus neutralization.
Deng, L., Hernandez, N., Zhong, L., Holcomb, D.D., Yan, H., Virata, M.L., Tarafdar, S., Xu, Y., He, Y., Struble, E., Alter, H.J., Zhang, P.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34260404
- DOI: https://doi.org/10.1073/pnas.2104242118
- Primary Citation of Related Structures:
7LKI - PubMed Abstract:
Epitope III, a highly conserved amino acid motif of 524 APTYSW 529 on the hepatitis C virus (HCV) E2 glycoprotein, resides in the critical loop that binds to the host receptor CD81, thus making it one of the most important antibody targets for blocking HCV infections. Here, we have determined the X-ray crystal structure of epitope III at a 2.0-Å resolution when it was captured by a site-specific neutralizing antibody, monoclonal antibody 1H8 (mAb1H8). The snapshot of this complex revealed that epitope III has a relatively rigid structure when confined in the binding grooves of mAb1H8, which confers the residue specificity at both ends of the epitope. Such a high shape complementarity is reminiscent of the "lock and key" mode of action, which is reinforced by the incompatibility of an antibody binding with an epitope bearing specific mutations. By subtly positioning the side chains on the three residues of Tyr 527 , Ser 528 , and Trp 529 while preserving the spatial rigidity of the rest, epitope III in this cocrystal complex adopts a unique conformation that is different from previously described E2 structures. With further analyses of molecular docking and phage display-based peptide interactions, we recognized that it is the arrangements of two separate sets of residues within epitope III that create these discrete conformations for the epitope to interact selectively with either mAb1H8 or CD81. These observations thus raise the possibility that local epitope III conformational dynamics, in conjunction with sequence variations, may act as a regulatory mechanism to coordinate "mAb1H8-like" antibody-mediated immune defenses with CD81-initiated HCV infections.
- Division of Plasma Protein Therapeutics, Office of Tissues and Advanced Therapies, Center for Biologics Evaluation and Research, Food and Drug Administration, Silver Spring, MD 20993-0002.
Organizational Affiliation: