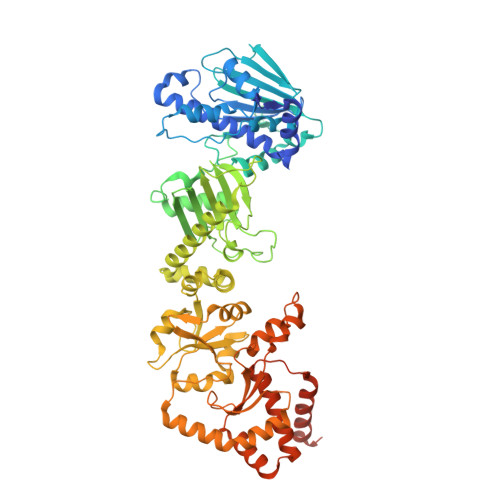
The structure of an Hsp90-immunophilin complex reveals cochaperone recognition of the client maturation state.
Lee, K., Thwin, A.C., Nadel, C.M., Tse, E., Gates, S.N., Gestwicki, J.E., Southworth, D.R.(2021) Mol Cell 81: 3496
- PubMed: 34380015
- DOI: https://doi.org/10.1016/j.molcel.2021.07.023
- Primary Citation of Related Structures:
7L7I, 7L7J - PubMed Abstract:
The Hsp90 chaperone promotes folding and activation of hundreds of client proteins in the cell through an ATP-dependent conformational cycle guided by distinct cochaperone regulators. The FKBP51 immunophilin binds Hsp90 with its tetratricopeptide repeat (TPR) domain and catalyzes peptidyl-prolyl isomerase (PPIase) activity during folding of kinases, nuclear receptors, and tau. Here we determined the cryoelectron microscopy (cryo-EM) structure of the human Hsp90:FKBP51:p23 complex to 3.3 Å, which, together with mutagenesis and crosslinking analyses, reveals the basis for cochaperone binding to Hsp90 during client maturation. A helix extension in the TPR functions as a key recognition element, interacting across the Hsp90 C-terminal dimer interface presented in the closed, ATP conformation. The PPIase domain is positioned along the middle domain, adjacent to Hsp90 client binding sites, whereas a single p23 makes stabilizing interactions with the N-terminal dimer. With this architecture, FKBP51 is positioned to act on specific client residues presented during Hsp90-catalyzed remodeling.
- Institute for Neurodegenerative Diseases, University of California, San Francisco, San Francisco, CA 94158, USA.
Organizational Affiliation: