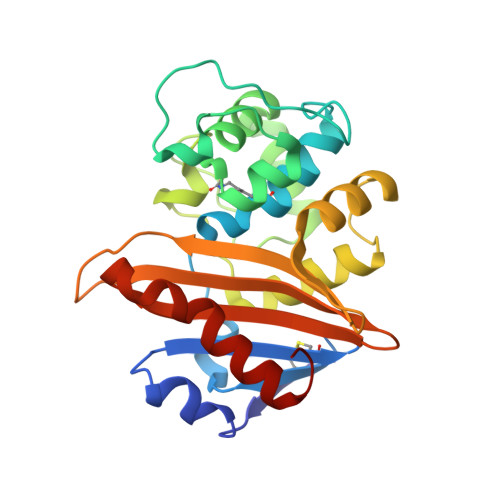
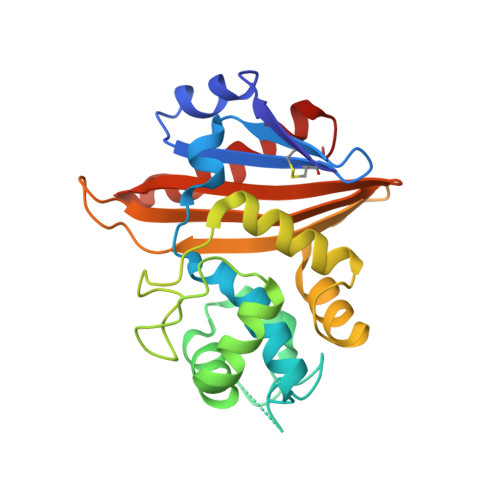
Functional and Structural Characterization of OXA-935, a Novel OXA-10-Family beta-Lactamase from Pseudomonas aeruginosa.
Pincus, N.B., Rosas-Lemus, M., Gatesy, S.W.M., Bertucci, H.K., Brunzelle, J.S., Minasov, G., Shuvalova, L.A., Lebrun-Corbin, M., Satchell, K.J.F., Ozer, E.A., Hauser, A.R., Bachta, K.E.R.(2022) Antimicrob Agents Chemother 66: e0098522-e0098522
- PubMed: 36129295
- DOI: https://doi.org/10.1128/aac.00985-22
- Primary Citation of Related Structures:
7L5R, 7L5V, 7N1M - PubMed Abstract:
Resistance to antipseudomonal penicillins and cephalosporins is often driven by the overproduction of the intrinsic β-lactamase AmpC. However, OXA-10-family β-lactamases are a rich source of resistance in Pseudomonas aeruginosa. OXA β-lactamases have a propensity for mutation that leads to extended spectrum cephalosporinase and carbapenemase activity. In this study, we identified isolates from a subclade of the multidrug-resistant (MDR) high risk P. aeruginosa clonal complex CC446 with a resistance to ceftazidime. A genomic analysis revealed that these isolates harbored a plasmid containing a novel allele of bla OXA-10 , named bla OXA-935 , which was predicted to produce an OXA-10 variant with two amino acid substitutions: an aspartic acid instead of a glycine at position 157 and a serine instead of a phenylalanine at position 153. The G157D mutation, present in OXA-14, is associated with the resistance of P. aeruginosa to ceftazidime. Compared to OXA-14, OXA-935 showed increased catalytic efficiency for ceftazidime. The deletion of bla OXA-935 restored the sensitivity to ceftazidime, and susceptibility profiling of P. aeruginosa laboratory strains expressing bla OXA-935 revealed that OXA-935 conferred ceftazidime resistance. To better understand the impacts of the variant amino acids, we determined the crystal structures of OXA-14 and OXA-935. Compared to OXA-14, the F153S mutation in OXA-935 conferred increased flexibility in the omega (Ω) loop. Amino acid changes that confer extended spectrum cephalosporinase activity to OXA-10-family β-lactamases are concerning, given the rising reliance on novel β-lactam/β-lactamase inhibitor combinations, such as ceftolozane-tazobactam and ceftazidime-avibactam, to treat MDR P. aeruginosa infections.
- Department of Microbiology-Immunology, Northwestern Universitygrid.16753.36, Feinberg School of Medicine, Chicago, Illinois, USA.
Organizational Affiliation: