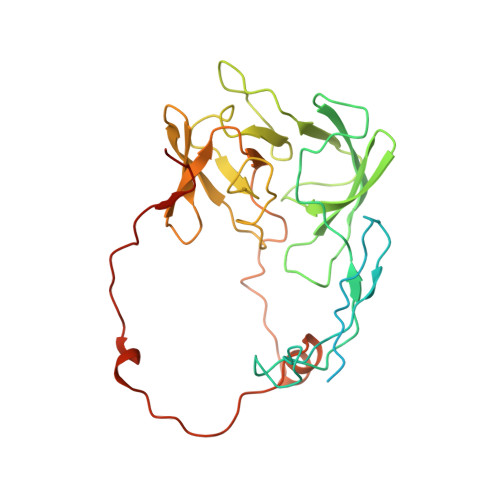
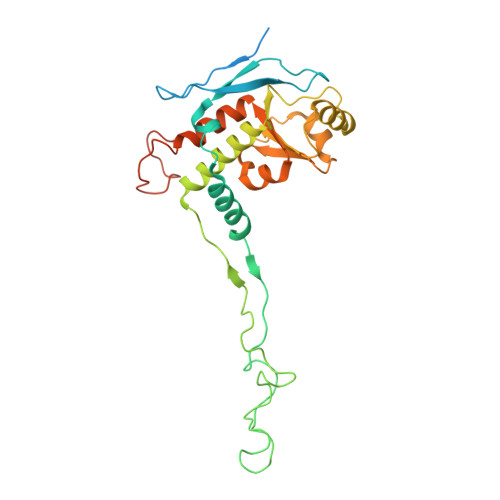
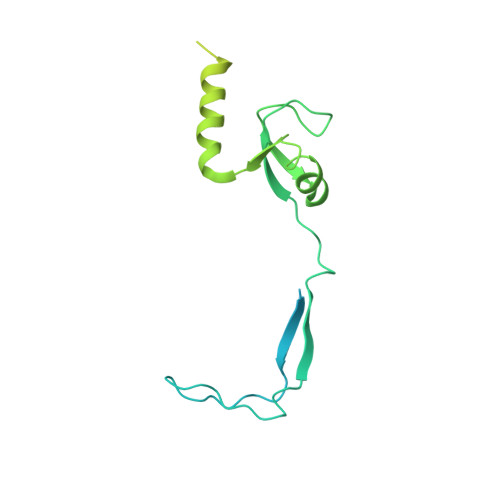
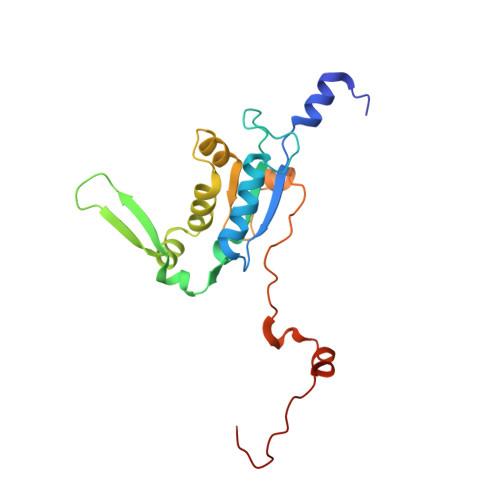
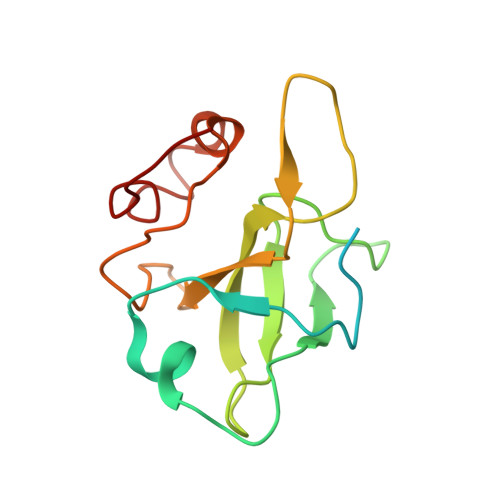
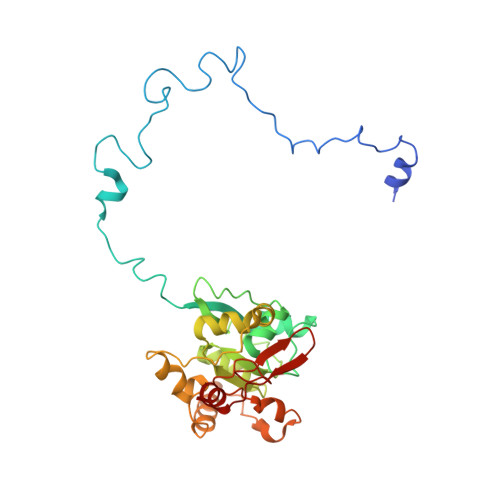
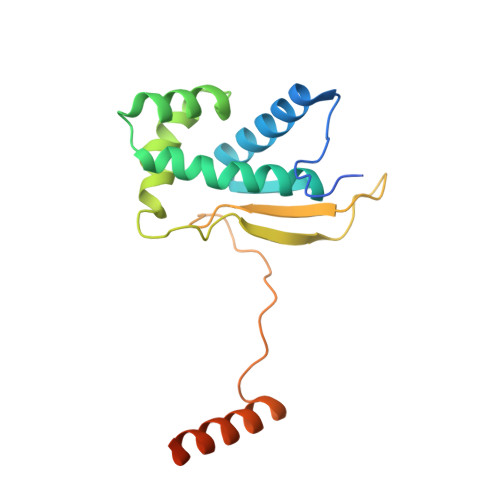
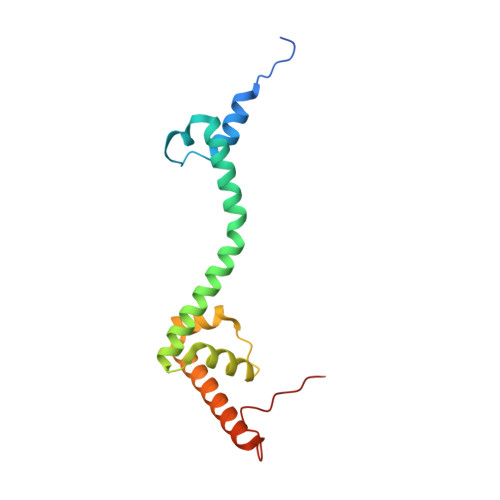
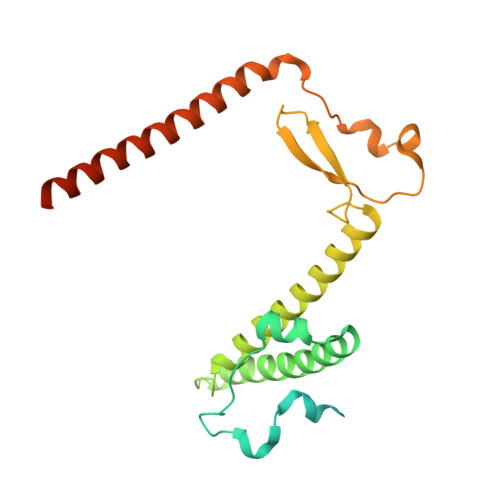
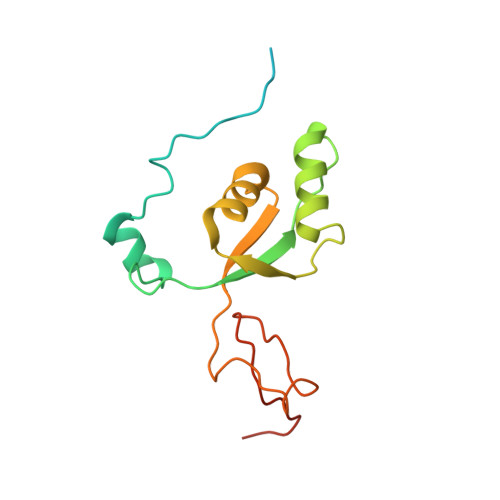
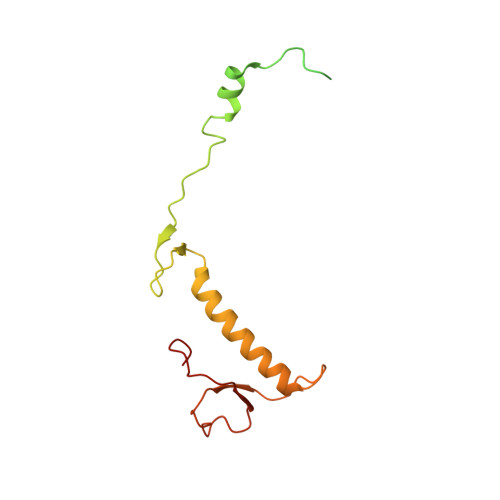
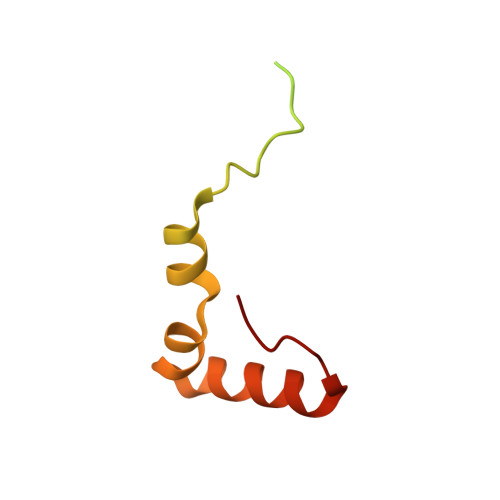
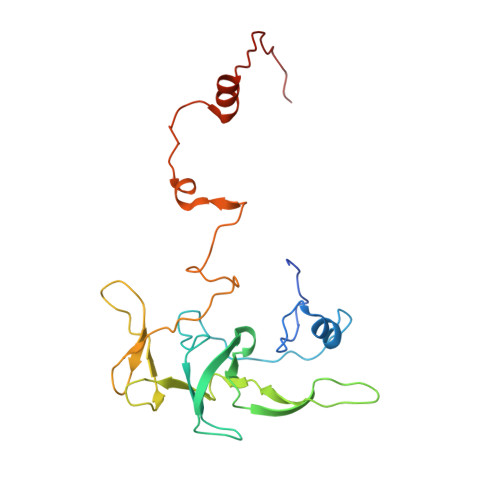
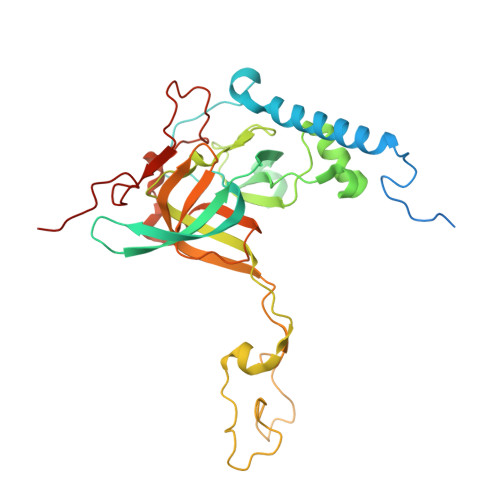
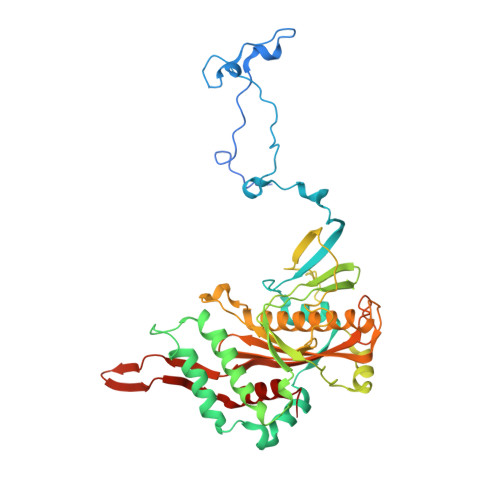
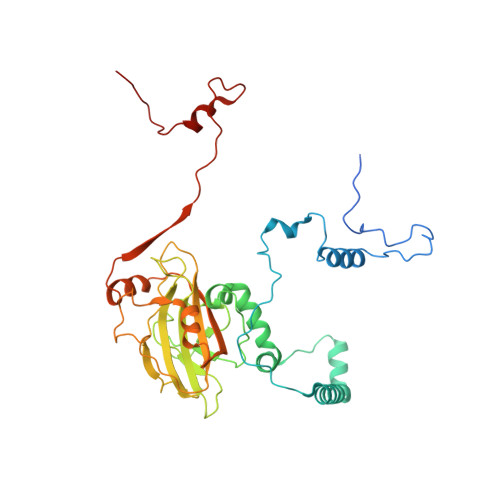
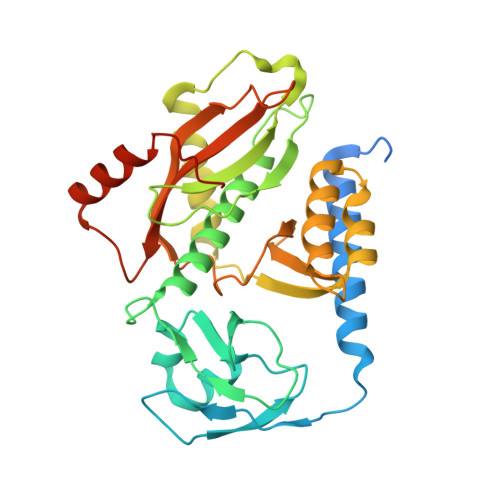
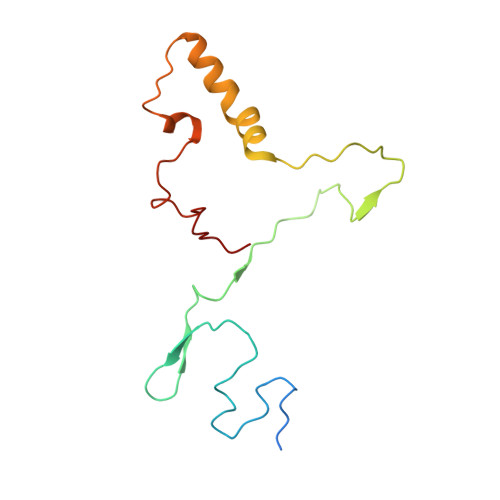
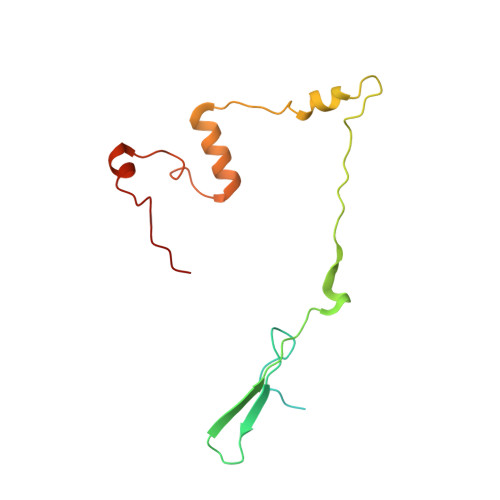
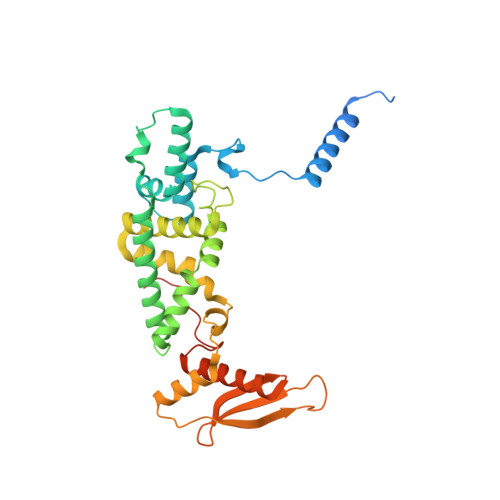
Distinct mechanisms of the human mitoribosome recycling and antibiotic resistance.
Koripella, R.K., Deep, A., Agrawal, E.K., Keshavan, P., Banavali, N.K., Agrawal, R.K.(2021) Nat Commun 12: 3607-3607
- PubMed: 34127662
- DOI: https://doi.org/10.1038/s41467-021-23726-4
- Primary Citation of Related Structures:
7L08, 7L20 - PubMed Abstract:
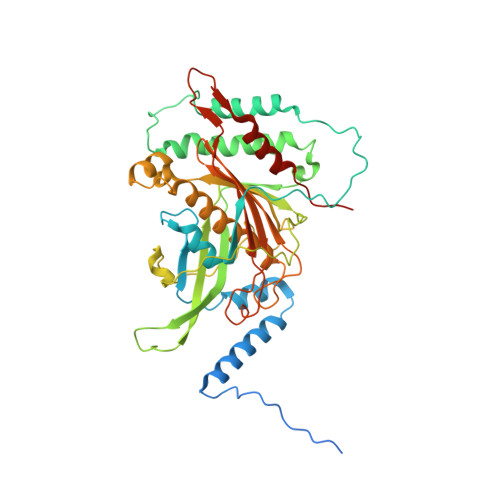
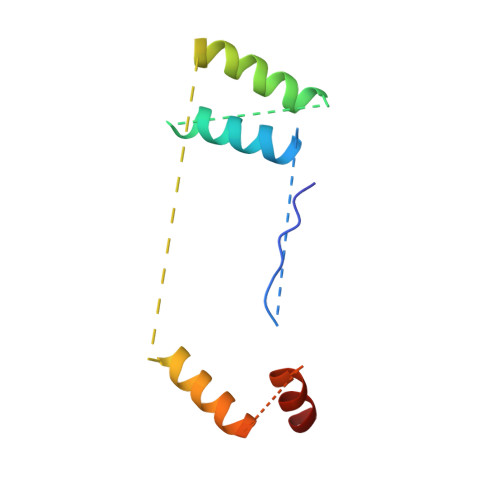
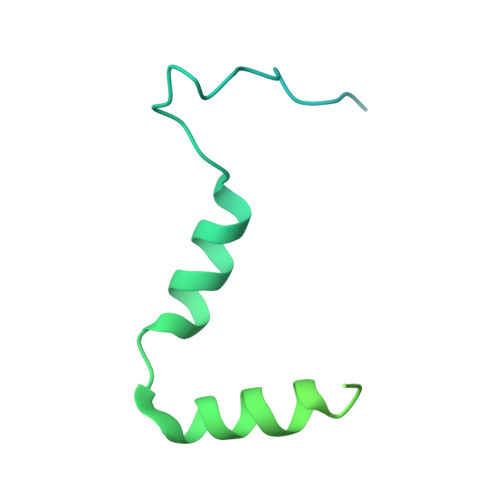
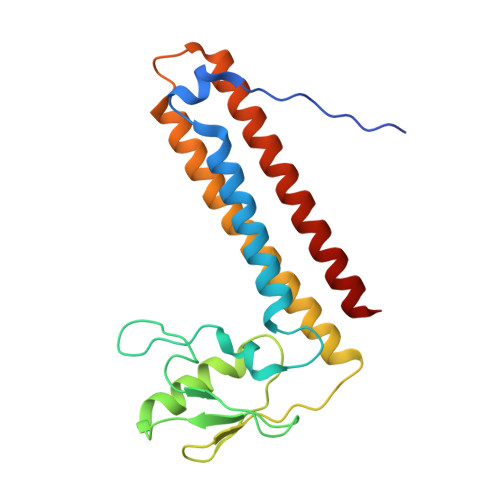
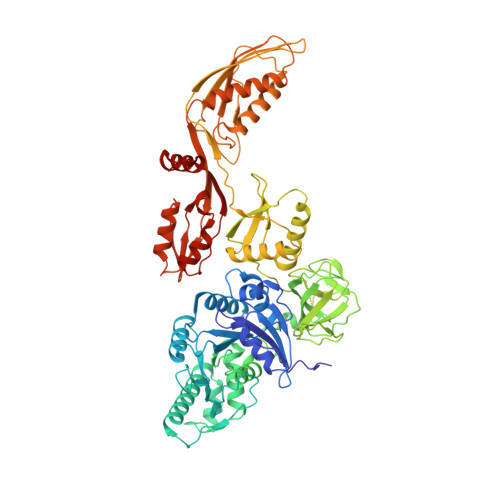
Ribosomes are recycled for a new round of translation initiation by dissociation of ribosomal subunits, messenger RNA and transfer RNA from their translational post-termination complex. Here we present cryo-EM structures of the human 55S mitochondrial ribosome (mitoribosome) and the mitoribosomal large 39S subunit in complex with mitoribosome recycling factor (RRF mt ) and a recycling-specific homolog of elongation factor G (EF-G2 mt ). These structures clarify an unusual role of a mitochondria-specific segment of RRF mt , identify the structural distinctions that confer functional specificity to EF-G2 mt , and show that the deacylated tRNA remains with the dissociated 39S subunit, suggesting a distinct sequence of events in mitoribosome recycling. Furthermore, biochemical and structural analyses reveal that the molecular mechanism of antibiotic fusidic acid resistance for EF-G2 mt is markedly different from that of mitochondrial elongation factor EF-G1 mt , suggesting that the two human EF-G mt s have evolved diversely to negate the effect of a bacterial antibiotic.
- Division of Translational Medicine, Wadsworth Center, New York State Department of Health, Empire State Plaza, Albany, NY, USA.
Organizational Affiliation: