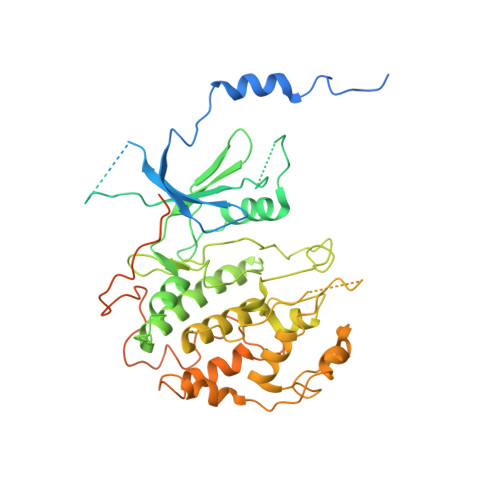
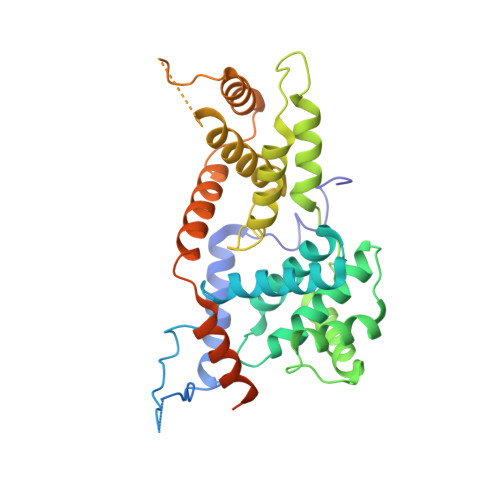
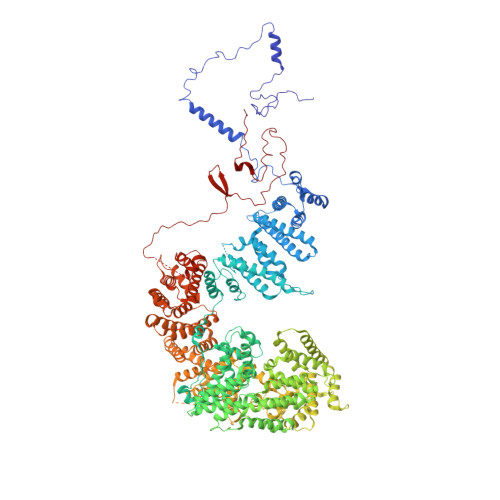
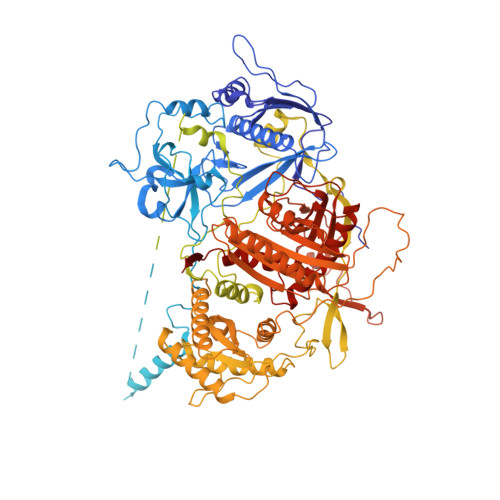
Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Li, Y.C., Chao, T.C., Kim, H.J., Cholko, T., Chen, S.F., Li, G., Snyder, L., Nakanishi, K., Chang, C.E., Murakami, K., Garcia, B.A., Boyer, T.G., Tsai, K.L.(2021) Sci Adv 7
- PubMed: 33523904
- DOI: https://doi.org/10.1126/sciadv.abd4484
- Primary Citation of Related Structures:
7KPV, 7KPW, 7KPX - PubMed Abstract:
The Cdk8 kinase module (CKM) in Mediator, comprising Med13, Med12, CycC, and Cdk8, regulates RNA polymerase II transcription through kinase-dependent and -independent functions. Numerous pathogenic mutations causative for neurodevelopmental disorders and cancer congregate in CKM subunits. However, the structure of the intact CKM and the mechanism by which Cdk8 is non-canonically activated and functionally affected by oncogenic CKM alterations are poorly understood. Here, we report a cryo-electron microscopy structure of Saccharomyces cerevisiae CKM that redefines prior CKM structural models and explains the mechanism of Med12-dependent Cdk8 activation. Med12 interacts extensively with CycC and activates Cdk8 by stabilizing its activation (T-)loop through conserved Med12 residues recurrently mutated in human tumors. Unexpectedly, Med13 has a characteristic Argonaute-like bi-lobal architecture. These findings not only provide a structural basis for understanding CKM function and pathological dysfunction, but also further impute a previously unknown regulatory mechanism of Mediator in transcriptional modulation through its Med13 Argonaute-like features.
- Department of Biochemistry and Molecular Biology, McGovern Medical School, University of Texas Health Science Center at Houston, Houston, TX, USA.
Organizational Affiliation: