K29-linked ubiquitin signaling regulates proteotoxic stress response and cell cycle.
Yu, Y., Zheng, Q., Erramilli, S.K., Pan, M., Park, S., Xie, Y., Li, J., Fei, J., Kossiakoff, A.A., Liu, L., Zhao, M.(2021) Nat Chem Biol 17: 896-905
- PubMed: 34239127
- DOI: https://doi.org/10.1038/s41589-021-00823-5
- Primary Citation of Related Structures:
7KEO - PubMed Abstract:
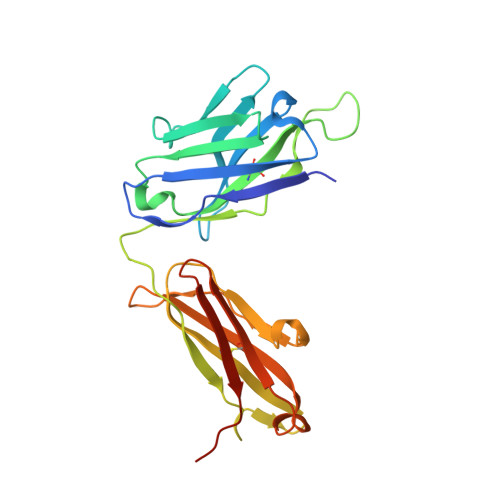
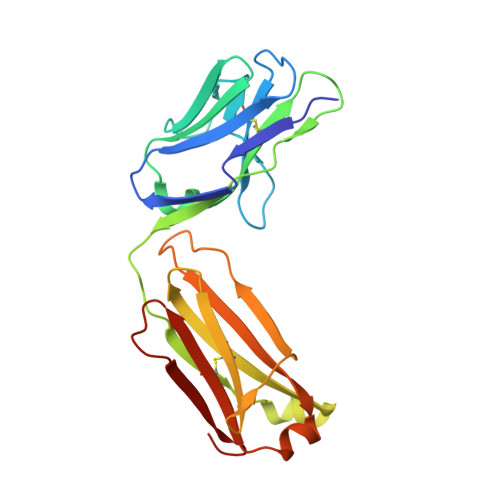
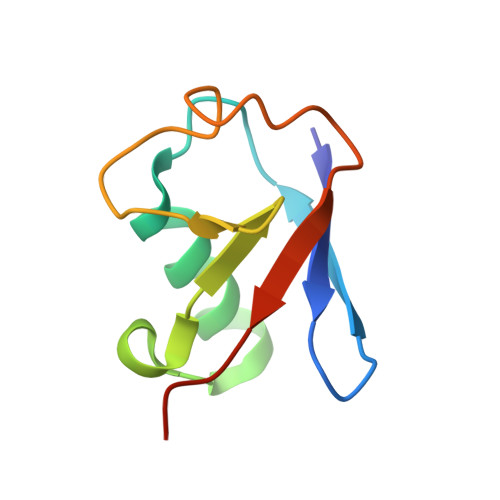
Protein ubiquitination shows remarkable topological and functional diversity through the polymerization of ubiquitin via different linkages. Deciphering the cellular ubiquitin code is of central importance to understand the physiology of the cell. However, our understanding of its function is rather limited due to the lack of specific binders as tools to detect K29-linked polyubiquitin. In this study, we screened and characterized a synthetic antigen-binding fragment, termed sAB-K29, that can specifically recognize K29-linked polyubiquitin using chemically synthesized K29-linked diubiquitin. We further determined the crystal structure of this fragment bound to the K29-linked diubiquitin, which revealed the molecular basis of specificity. Using sAB-K29 as a tool, we uncovered that K29-linked ubiquitination is involved in different kinds of cellular proteotoxic stress response as well as cell cycle regulation. In particular, we showed that K29-linked ubiquitination is enriched in the midbody and downregulation of the K29-linked ubiquitination signal arrests cells in G1/S phase.
- Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL, USA.
Organizational Affiliation: