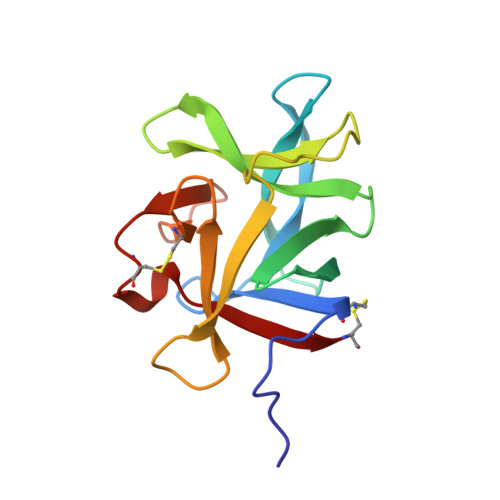
The structures of two salivary proteins from the West Nile vector Culex quinquefasciatus reveal a beta-trefoil fold with putative sugar binding properties
Kern, O., Valenzuela Leon, P.C., Gittis, A.G., Bonilla, B., Cruz, P., Chagas, A.C., Ganesan, S., Ribeiro, J.M., Garboczi, D.N., Martin-Martin, I., Calvo, E.(2021) Curr Res Struct Biol 3: 95-105