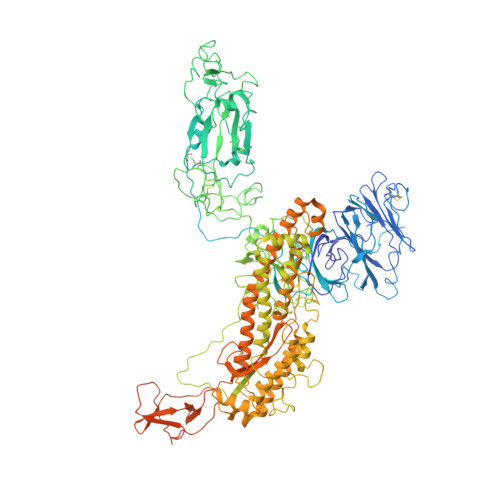
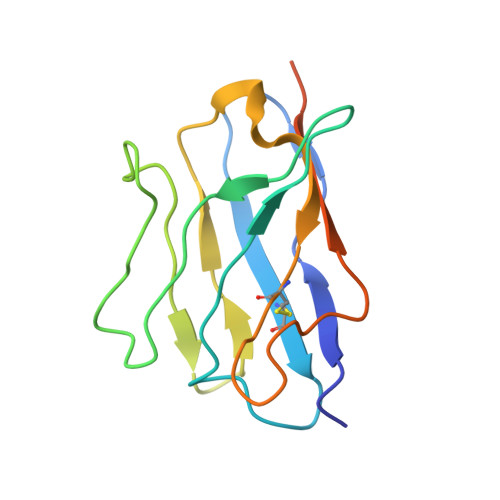
Nasal delivery of single-domain antibody improves symptoms of SARS-CoV-2 infection in an animal model.
Haga, K., Takai-Todaka, R., Matsumura, Y., Song, C., Takano, T., Tojo, T., Nagami, A., Ishida, Y., Masaki, H., Tsuchiya, M., Ebisudani, T., Sugimoto, S., Sato, T., Yasuda, H., Fukunaga, K., Sawada, A., Nemoto, N., Murata, K., Morimoto, T., Katayama, K.(2021) PLoS Pathog 17: e1009542-e1009542
- PubMed: 34648602
- DOI: https://doi.org/10.1371/journal.ppat.1009542
- Primary Citation of Related Structures:
7FG2, 7FG3, 7FG7 - PubMed Abstract:
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that causes the disease COVID-19 can lead to serious symptoms, such as severe pneumonia, in the elderly and those with underlying medical conditions. While vaccines are now available, they do not work for everyone and therapeutic drugs are still needed, particularly for treating life-threatening conditions. Here, we showed nasal delivery of a new, unmodified camelid single-domain antibody (VHH), termed K-874A, effectively inhibited SARS-CoV-2 titers in infected lungs of Syrian hamsters without causing weight loss and cytokine induction. In vitro studies demonstrated that K-874A neutralized SARS-CoV-2 in both VeroE6/TMPRSS2 and human lung-derived alveolar organoid cells. Unlike other drug candidates, K-874A blocks viral membrane fusion rather than viral attachment. Cryo-electron microscopy revealed K-874A bound between the receptor binding domain and N-terminal domain of the virus S protein. Further, infected cells treated with K-874A produced fewer virus progeny that were less infective. We propose that direct administration of K-874A to the lung could be a new treatment for preventing the reinfection of amplified virus in COVID-19 patients.
- Laboratory of Viral Infection, Department of Infection Control and Immunology, Ōmura Satoshi Memorial Institute & Graduate School of Infection Control Sciences, Kitasato University, Tokyo, Japan.
Organizational Affiliation: