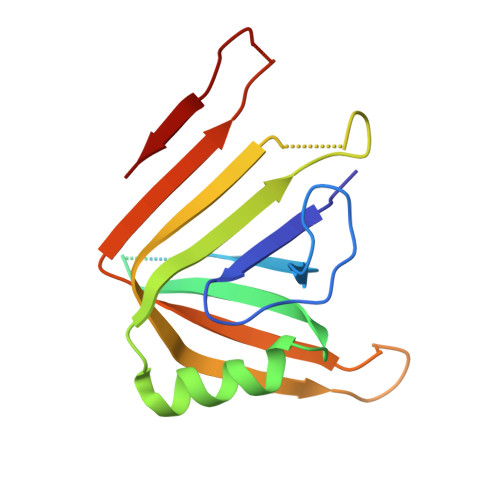
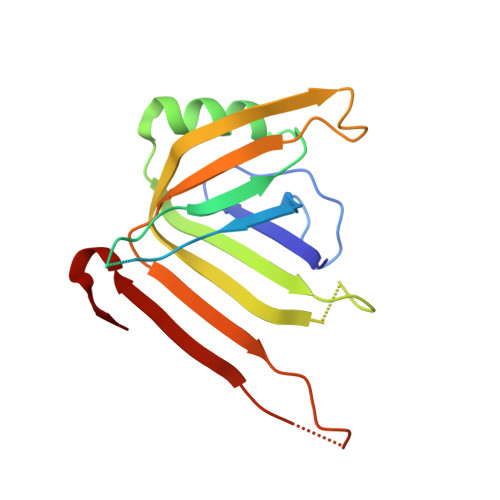
Bacterial antagonism of Chromobacterium haemolyticum and characterization of its putative type VI secretion system.
Zwe, Y.H., Yadav, M., Ten, M.M.Z., Srinivasan, M., Jobichen, C., Sivaraman, J., Li, D.(2022) Res Microbiol 173: 103918-103918
- PubMed: 34906677
- DOI: https://doi.org/10.1016/j.resmic.2021.103918
- Primary Citation of Related Structures:
7FCF - PubMed Abstract:
This study reports the isolation of a new Chromobacterium haemolyticum strain named WI5 from a hydroponic farming facility. WI5 exhibited remarkable bacterial antagonistic properties, eliminating Salmonella, Escherichia coli, Listeria monocytogenes and Staphylococcus aureus (initial inoculum load ∼10 5 CFU/ml) in dual-species co-culture biofilms. Antagonism was strictly contact-dependent and highly influenced by nutrient availability. Next, we identified a complete suite of putative Type VI secretion system (T6SS) genes in the WI5 genome, annotated the gene locus architecture, and determined the crystal structure of hallmark T6SS tube protein Hcp1, which revealed a hexameric ring structure with an outer and inner diameter of 77 and 45 Å, respectively. Structural comparison with homologs showed differences in the key loops connecting the β-strands in which the conserved residues are located, suggesting a role of these residues in the protein function. The T6SS is well-known to facilitate interbacterial competition, and the putative T6SS characterized herein might be responsible for the remarkable antagonism by C. haemolyticum WI5. Collectively, these findings shed light on the nature of bacterial antagonism and a putative key virulence determinant of C. haemolyticum, which might aid in further understanding its potential ecological role in natural habitats.
- Department of Food Science & Technology, 2 Science Drive 2, Faculty of Science, National University of Singapore, Singapore, 117543.
Organizational Affiliation: