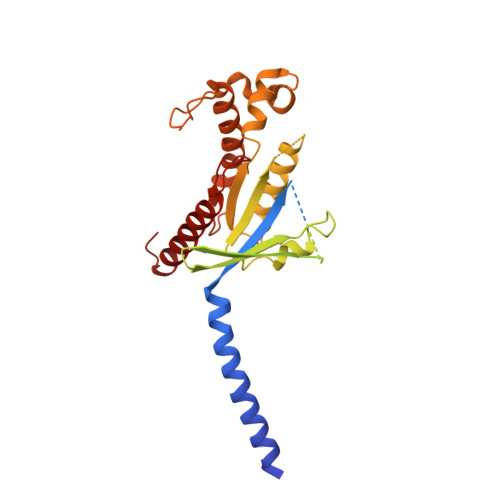
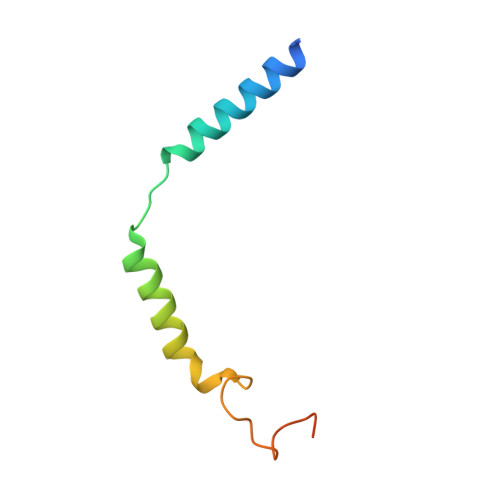
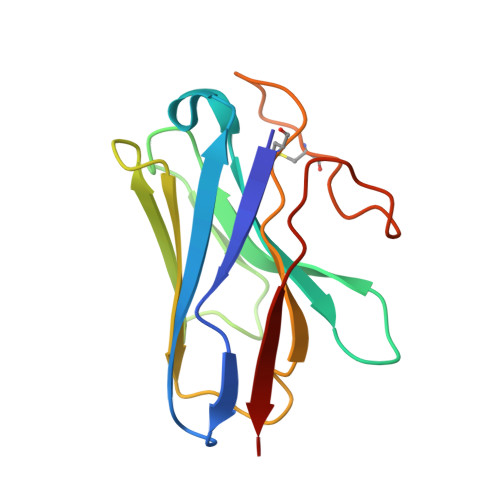
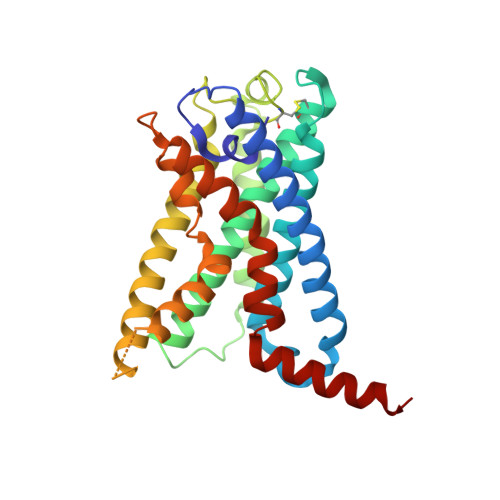
Structural basis for the tethered peptide activation of adhesion GPCRs.
Ping, Y.Q., Xiao, P., Yang, F., Zhao, R.J., Guo, S.C., Yan, X., Wu, X., Zhang, C., Lu, Y., Zhao, F., Zhou, F., Xi, Y.T., Yin, W., Liu, F.Z., He, D.F., Zhang, D.L., Zhu, Z.L., Jiang, Y., Du, L., Feng, S.Q., Schoneberg, T., Liebscher, I., Xu, H.E., Sun, J.P.(2022) Nature 604: 763-770
- PubMed: 35418678
- DOI: https://doi.org/10.1038/s41586-022-04619-y
- Primary Citation of Related Structures:
7EPT, 7EQ1 - PubMed Abstract:
Adhesion G-protein-coupled receptors (aGPCRs) are important for organogenesis, neurodevelopment, reproduction and other processes 1-6 . Many aGPCRs are activated by a conserved internal (tethered) agonist sequence known as the Stachel sequence 7-12 . Here, we report the cryogenic electron microscopy (cryo-EM) structures of two aGPCRs in complex with G s : GPR133 and GPR114. The structures indicate that the Stachel sequences of both receptors assume an α-helical-bulge-β-sheet structure and insert into a binding site formed by the transmembrane domain (TMD). A hydrophobic interaction motif (HIM) within the Stachel sequence mediates most of the intramolecular interactions with the TMD. Combined with the cryo-EM structures, biochemical characterization of the HIM motif provides insight into the cross-reactivity and selectivity of the Stachel sequences. Two interconnected mechanisms, the sensing of Stachel sequences by the conserved 'toggle switch' W 6.53 and the constitution of a hydrogen-bond network formed by Q 7.49 /Y 7.49 and the P 6.47 /V 6.47 φφG 6.50 motif (φ indicates a hydrophobic residue), are important in Stachel sequence-mediated receptor activation and G s coupling. Notably, this network stabilizes kink formation in TM helices 6 and 7 (TM6 and TM7, respectively). A common G s -binding interface is observed between the two aGPCRs, and GPR114 has an extended TM7 that forms unique interactions with G s . Our structures reveal the detailed mechanisms of aGPCR activation by Stachel sequences and their G s coupling.
- Key Laboratory Experimental Teratology of the Ministry of Education, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Cheeloo College of Medicine, Shandong University, Jinan, China.
Organizational Affiliation: