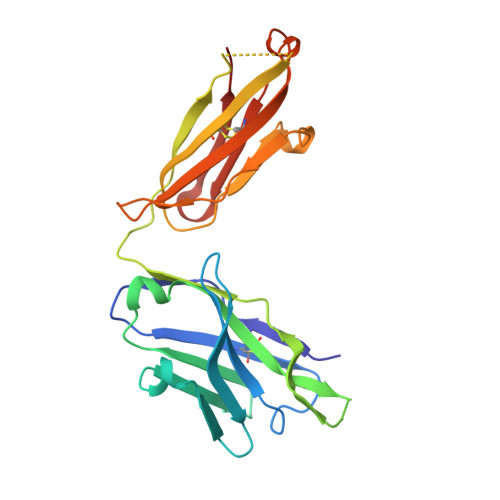
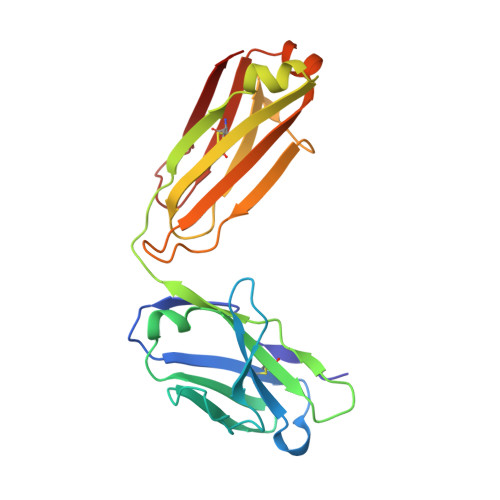
Focal accumulation of aromaticity at the CDRH3 loop mitigates 4E10 polyreactivity without altering its HIV neutralization profile.
Rujas, E., Leaman, D.P., Insausti, S., Carravilla, P., Garcia-Porras, M., Largo, E., Morillo, I., Sanchez-Eugenia, R., Zhang, L., Cui, H., Iloro, I., Elortza, F., Julien, J.P., Eggeling, C., Zwick, M.B., Caaveiro, J.M.M., Nieva, J.L.(2021) iScience 24: 102987-102987
- PubMed: 34505005
- DOI: https://doi.org/10.1016/j.isci.2021.102987
- Primary Citation of Related Structures:
7EKB, 7EKK - PubMed Abstract:
Broadly neutralizing antibodies (bnAbs) against HIV-1 are frequently associated with the presence of autoreactivity/polyreactivity, a property that can limit their use as therapeutic agents. The bnAb 4E10, targeting the conserved Membrane proximal external region (MPER) of HIV-1, displays almost pan-neutralizing activity across globally circulating HIV-1 strains but exhibits nonspecific off-target interactions with lipid membranes. The hydrophobic apex of the third complementarity-determining region of the heavy chain (CDRH3) loop, which is essential for viral neutralization, critically contributes to this detrimental effect. Here, we have replaced the aromatic/hydrophobic residues from the apex of the CDRH3 of 4E10 with a single aromatic molecule through chemical modification to generate a variant that preserves the neutralization potency and breadth of 4E10 but with reduced autoreactivity. Collectively, our study suggests that the localized accumulation of aromaticity by chemical modification provides a pathway to ameliorate the adverse effects triggered by the CDRH3 of anti-HIV-1 MPER bnAbs.
- Instituto Biofisika (CSIC, UPV/EHU) and Department of Biochemistry and Molecular Biology, University of the Basque Country (UPV/EHU), P.O. Box 644, 48080 Bilbao, Spain.
Organizational Affiliation: