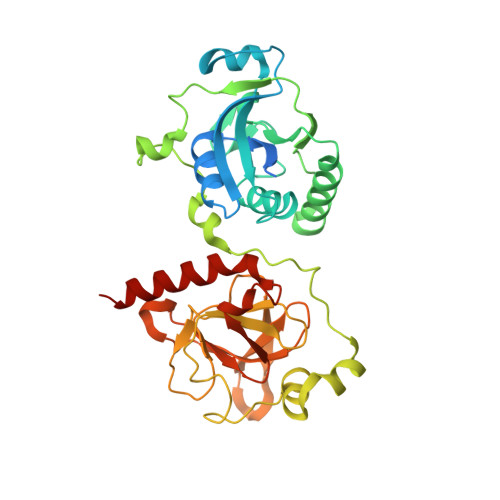
Structural Insights into a Bifunctional Peptide Methionine Sulfoxide Reductase MsrA/B Fusion Protein from Helicobacter pylori .
Kim, S., Lee, K., Park, S.H., Kwak, G.H., Kim, M.S., Kim, H.Y., Hwang, K.Y.(2021) Antioxidants (Basel) 10
- PubMed: 33807684
- DOI: https://doi.org/10.3390/antiox10030389
- Primary Citation of Related Structures:
7E43 - PubMed Abstract:
Methionine sulfoxide reductase (Msr) is a family of enzymes that reduces oxidized methionine and plays an important role in the survival of bacteria under oxidative stress conditions. MsrA and MsrB exist in a fusion protein form (MsrAB) in some pathogenic bacteria, such as Helicobacter pylori ( Hp ), Streptococcus pneumoniae , and Treponema denticola . To understand the fused form instead of the separated enzyme at the molecular level, we determined the crystal structure of Hp MsrAB C44S/C318S at 2.2 Å, which showed that a linker region ( Hpiloop , 193-205) between two domains interacted with each Hp MsrA or Hp MsrB domain via three salt bridges (E193-K107, D197-R103, and K200-D339). Two acetate molecules in the active site pocket showed an sp 2 planar electron density map in the crystal structure, which interacted with the conserved residues in fusion MsrABs from the pathogen. Biochemical and kinetic analyses revealed that Hpiloop is required to increase the catalytic efficiency of Hp MsrAB. Two salt bridge mutants (D193A and E199A) were located at the entrance or tailgate of Hpiloop . Therefore, the linker region of the MsrAB fusion enzyme plays a key role in the structural stability and catalytic efficiency and provides a better understanding of why MsrAB exists in a fused form.
- Department of Biotechnology, Korea University, Seoul 02841, Korea.
Organizational Affiliation: