Crystal structure and cellular functions of uPAR dimer
Yu, S., Sui, Y., Wang, J., Li, Y., Li, H., Cao, Y., Chen, L., Jiang, L., Yuan, C., Huang, M.(2022) Nat Commun 13: 1665
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Urokinase plasminogen activator surface receptor | 279 | Homo sapiens | Mutation(s): 0 Gene Names: PLAUR, MO3, UPAR | 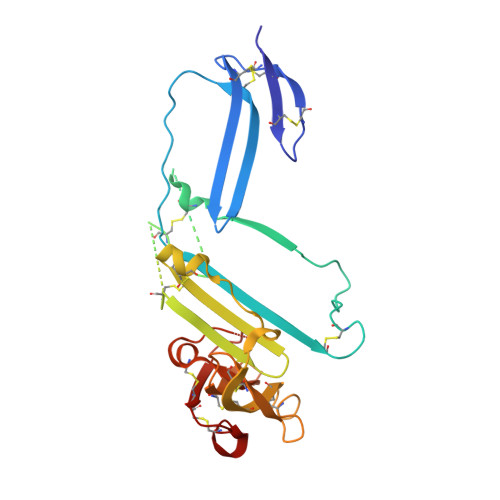 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q03405 (Homo sapiens) Explore Q03405 Go to UniProtKB: Q03405 | |||||
PHAROS: Q03405 GTEx: ENSG00000011422 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q03405 | ||||
Glycosylation | |||||
| Glycosylation Sites: 3 | Go to GlyGen: Q03405-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG (Subject of Investigation/LOI) Query on NAG | C [auth A] D [auth A] E [auth A] F [auth B] G [auth B] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 79.58 | α = 90 |
| b = 79.58 | β = 90 |
| c = 270.53 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| xia2 | data reduction |
| PDB_EXTRACT | data extraction |
| xia2 | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 22077016 |