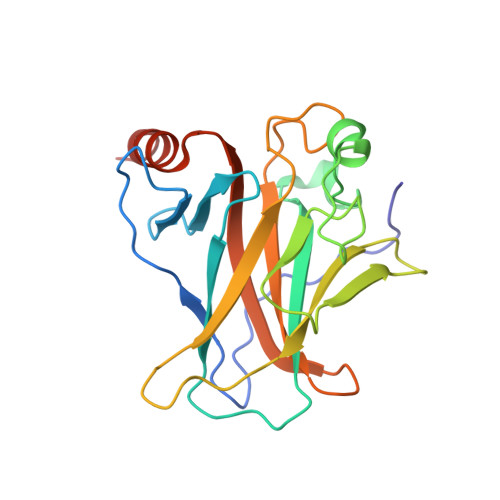
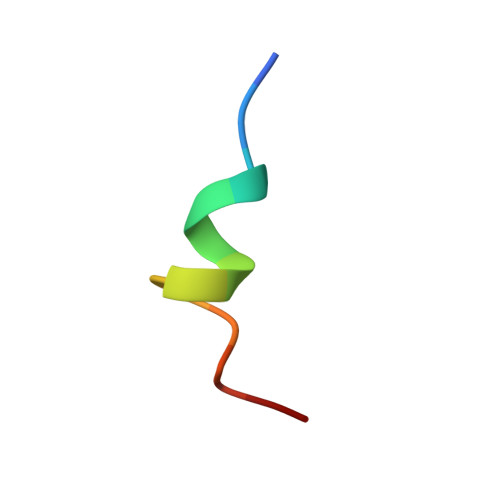
Structural basis of the p53 DNA binding domain and PUMA complex.
Han, C.W., Lee, H.N., Jeong, M.S., Park, S.Y., Jang, S.B.(2021) Biochem Biophys Res Commun 548: 39-46
- PubMed: 33631672
- DOI: https://doi.org/10.1016/j.bbrc.2021.02.049
- Primary Citation of Related Structures:
7DVD - PubMed Abstract:
PUMA (p53-upregulated modulator of apoptosis) is localized in mitochondria and a direct target in p53-mediated apoptosis. p53 elicits mitochondrial apoptosis via transcription-dependent and independent mechanisms. p53 is known to induce apoptosis via the transcriptional induction of PUMA, which encodes proapoptotic BH3-only members of the Bcl-2 protein family. However, the transcription-independent mechanisms of human PUMA remain poorly defined. For example, it is not known whether PUMA interacts directly with the DNA binding domain (DBD: residues 92-293) of p53 in vitro. Here, the structure of the complex between the DBD of p53 and PUMA peptide was elucidated by X-ray crystallography. Isothermal titration calorimetry showed that PUMA peptide binds strongly with p53 DBD, and the crystal structure of p53-PUMA peptide complex revealed it contains four molecules of p53 DBD and one PUMA peptide per asymmetric unit in space group P 1 . PUMA peptide bound to the N-terminal residues of p53 DBD. A cell proliferation assay demonstrated PUMA peptide inhibited the growth of a lung cancer cell line. These results contribute to understanding of the mechanism responsible for p53-mediated apoptosis.
- Department of Molecular Biology, College of Natural Sciences, Pusan National University, 2Busandaehak-ro 63beon-gil, Geumjeong-gu, Busan, 46241, Republic of Korea.
Organizational Affiliation: