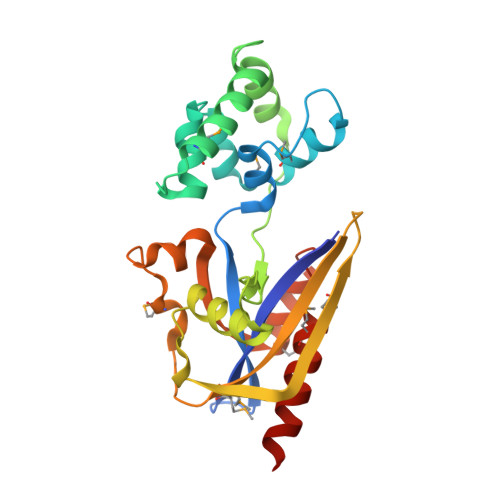
Structural Insight into Molecular Inhibitory Mechanism of InsP 6 on African Swine Fever Virus mRNA-Decapping Enzyme g5Rp.
Yang, Y., Zhang, C., Li, X., Li, L., Chen, Y., Yang, X., Zhao, Y., Chen, C., Wang, W., Zhong, Z., Yang, C., Huang, Z., Su, D.(2022) J Virol 96: e0190521-e0190521
- PubMed: 35481780
- DOI: https://doi.org/10.1128/jvi.01905-21
- Primary Citation of Related Structures:
7DNT, 7DNU - PubMed Abstract:
Removal of 5' cap on cellular mRNAs by the African swine fever virus (ASFV) decapping enzyme g5R protein (g5Rp) is beneficial to viral gene expression during the early stages of infection. As the only nucleoside diphosphate-linked moiety X (Nudix) decapping enzyme encoded in the ASFV genome, g5Rp works in both the degradation of cellular mRNA and the hydrolyzation of the diphosphoinositol polyphosphates. Here, we report the structures of dimeric g5Rp and its complex with inositol hexakisphosphate (InsP 6 ). The two g5Rp protomers interact head to head to form a dimer, and the dimeric interface is formed by extensive polar and nonpolar interactions. Each protomer is composed of a unique N-terminal helical domain and a C-terminal classic Nudix domain. As g5Rp is an mRNA-decapping enzyme, we identified key residues, including K 8 , K 94 , K 95 , K 98 , K 175 , R 221 , and K 243 located on the substrate RNA binding interfaces of g5Rp which are important to RNA binding and decapping enzyme activity. Furthermore, the g5Rp-mediated mRNA decapping was inhibited by InsP 6 . The g5Rp-InsP 6 complex structure showed that the InsP 6 molecules occupy the same regions that primarily mediate g5Rp-RNA interaction, elucidating the roles of InsP 6 in the regulation of the viral decapping activity of g5Rp in mRNA degradation. Collectively, these results provide the structural basis of interaction between RNA and g5Rp and highlight the inhibitory mechanism of InsP 6 on mRNA decapping by g5Rp. IMPORTANCE ASF is a highly contagious hemorrhagic viral disease in domestic pigs which causes high mortality. Currently, there are still no effective vaccines or specific drugs available against this particular virus. The protein g5Rp is the only viral mRNA-decapping enzyme, playing an essential role in the machinery assembly of mRNA regulation and translation initiation. In this study, we solved the crystal structures of g5Rp dimer and complex with InsP 6 . Structure-based mutagenesis studies revealed critical residues involved in a candidate RNA binding region, which also play pivotal roles in complex with InsP 6 . Notably, InsP 6 can inhibit g5Rp activity by competitively blocking the binding of substrate mRNA to the enzyme. Our structure-function studies provide the basis for potential anti-ASFV inhibitor designs targeting the critical enzyme.
- State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan Universitygrid.13291.38 and Collaborative Innovation Center of Biotherapy, Chengdu, Sichuan, People's Republic of China.
Organizational Affiliation: