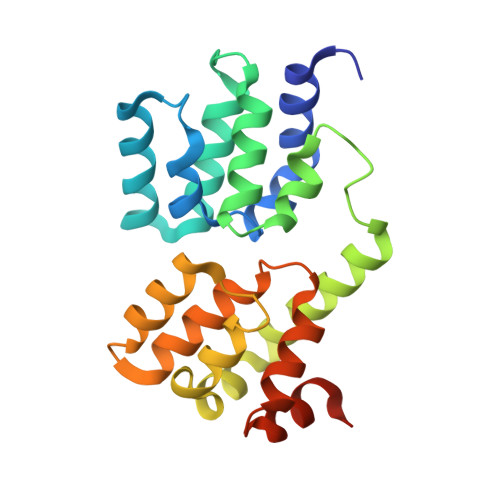
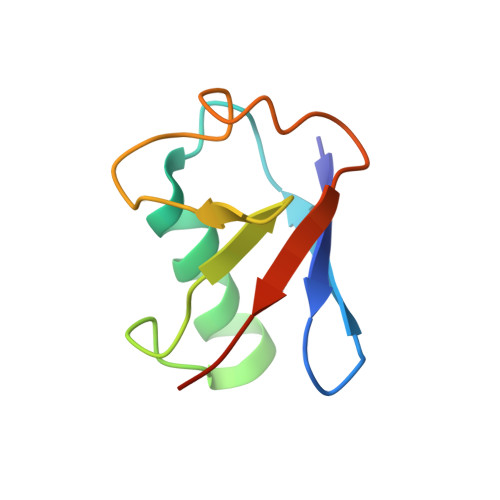
Ordered assembly of the cytosolic RNA-sensing MDA5-MAVS signaling complex via binding to unanchored K63-linked poly-ubiquitin chains.
Song, B., Chen, Y., Liu, X., Yuan, F., Tan, E.Y.J., Lei, Y., Song, N., Han, Y., Pascal, B.D., Griffin, P.R., Luo, C., Wu, B., Luo, D., Zheng, J.(2021) Immunity 54: 2218
- PubMed: 34644557
- DOI: https://doi.org/10.1016/j.immuni.2021.09.008
- Primary Citation of Related Structures:
7DNI, 7DNJ - PubMed Abstract:
The RNA sensor MDA5 recruits the signaling adaptor MAVS to initiate type I interferon signaling and downstream antiviral responses, a process that requires K63-linked polyubiquitin chains. Here, we examined the mechanisms whereby K63-polyUb chain regulate MDA5 activation. Only long unanchored K63-polyUb n (n ≥ 8) could mediate tetramerization of the caspase activation and recruitment domains of MDA5 ( MDA5 CARDs). Cryoelectron microscopy structures of a polyUb 13 -bound MDA5 CARDs tetramer and a polyUb 11 -bound MDA5 CARDs- MAVS CARD assembly revealed a tower-like formation, wherein eight Ubs tethered along the outer rim of the helical shell, bridging MDA5 CARDs and MAVS CARD tetramers into proximity. ATP binding and hydrolysis promoted the stabilization of RNA-bound MDA5 prior to MAVS activation via allosteric effects on CARDs-polyUb complex. Abundant ATP prevented basal activation of apo MDA5. Our findings reveal the ordered assembly of a MDA5 signaling complex competent to recruit and activate MAVS and highlight differences with RIG-I in terms of CARD orientation and Ub sensing that suggest different abilities to induce antiviral responses.
- Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China.
Organizational Affiliation: